mirror of https://github.com/mpastell/Weave.jl
Update tests
parent
ebd4f6dbed
commit
45a3f46918
13
.travis.yml
13
.travis.yml
|
|
@ -1,7 +1,6 @@
|
|||
|
||||
language: julia
|
||||
julia:
|
||||
- 0.5
|
||||
- 0.6
|
||||
- nightly
|
||||
matrix:
|
||||
|
|
@ -12,12 +11,12 @@ notifications:
|
|||
before_install:
|
||||
#- sudo apt-get update -qq -y
|
||||
#- sudo apt-get install python-matplotlib -y
|
||||
- wget http://repo.continuum.io/miniconda/Miniconda-latest-Linux-x86_64.sh -O miniconda.sh
|
||||
- chmod +x miniconda.sh
|
||||
- ./miniconda.sh -b
|
||||
- export PATH=/home/travis/miniconda2/bin:$PATH
|
||||
- conda update --yes conda
|
||||
- conda install --yes matplotlib
|
||||
#- wget http://repo.continuum.io/miniconda/Miniconda-latest-Linux-x86_64.sh -O miniconda.sh
|
||||
#- chmod +x miniconda.sh
|
||||
#- ./miniconda.sh -b
|
||||
#- export PATH=/home/travis/miniconda2/bin:$PATH
|
||||
#- conda update --yes conda
|
||||
#- conda install --yes matplotlib
|
||||
script:
|
||||
- if [[ -a .git/shallow ]]; then git fetch --unshallow; fi
|
||||
- julia --check-bounds=yes -e 'Pkg.clone(pwd()); Pkg.build("Weave")'
|
||||
|
|
|
|||
|
|
@ -26,10 +26,6 @@ and figures.
|
|||
|
||||
**Citing Weave:** *Pastell, Matti. 2017. Weave.jl: Scientific Reports Using Julia. The Journal of Open Source Software. http://dx.doi.org/10.21105/joss.00204*
|
||||
|
||||
|
||||
**Note about Julia 0.6**: Weave has been updated to support 0.6. PyPlot,
|
||||
Gadfly and Plots now all work, tests still need to be updated (15th May 2017).
|
||||
|
||||
|
||||
|
||||
## Installation
|
||||
|
|
|
|||
|
|
@ -1,7 +1,5 @@
|
|||
julia 0.5
|
||||
julia 0.6
|
||||
Cairo
|
||||
Gadfly
|
||||
PyPlot
|
||||
GR
|
||||
DSP
|
||||
Plots
|
||||
|
|
|
|||
|
|
@ -7,21 +7,22 @@ println(x)
|
|||
|
||||
|
||||
~~~~
|
||||
linspace(0.0,6.283185307179586,50)
|
||||
0.0:0.1282282715750936:6.283185307179586
|
||||
~~~~
|
||||
|
||||
|
||||
|
||||
~~~~{.julia}
|
||||
p = plot(x, sin(x), size =(900,300))
|
||||
p = plot(x, sin.(x), size =(900,300))
|
||||
p
|
||||
~~~~~~~~~~~~~
|
||||
|
||||
|
||||
\
|
||||
|
||||
|
||||
~~~~{.julia}
|
||||
julia> plot(x, sin(x))
|
||||
|
||||
|
||||
plot(x, sin.(x))
|
||||
~~~~~~~~~~~~~
|
||||
|
||||
|
||||
|
|
@ -37,24 +38,4 @@ scatter!(rand(100),markersize=6,c=:orange)
|
|||
\
|
||||
|
||||
|
||||
~~~~{.julia}
|
||||
julia> plot(rand(100) / 3,reg=true,fill=(0,:green))
|
||||
|
||||
|
||||
~~~~~~~~~~~~~
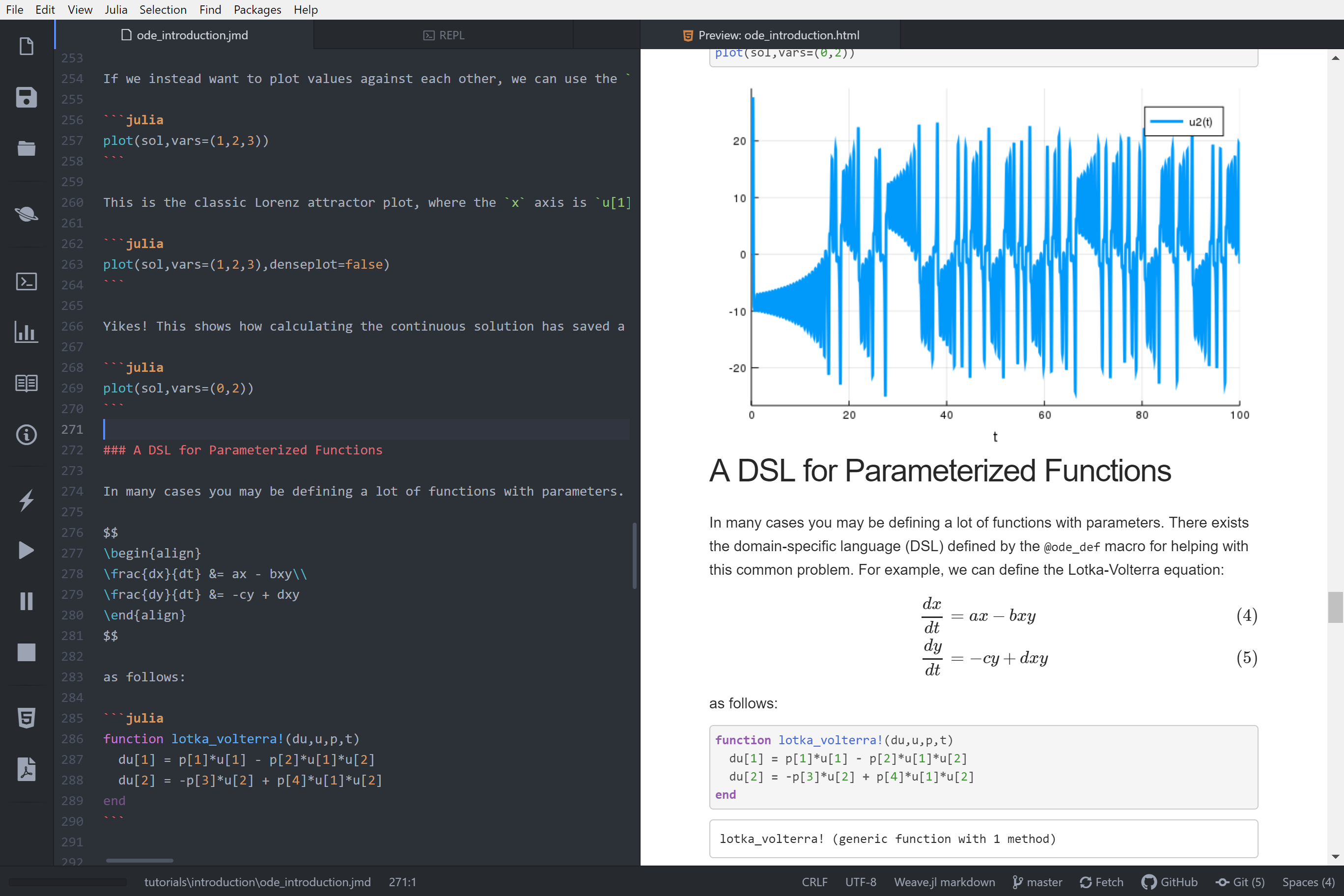
|
||||
|
||||
|
||||
\
|
||||
|
||||
|
||||
~~~~{.julia}
|
||||
julia> scatter!(rand(100),markersize=6,c=:orange)
|
||||
|
||||
|
||||
~~~~~~~~~~~~~
|
||||
|
||||
|
||||
\
|
||||
|
||||
|
||||

|
||||
|
|
|
|||
|
|
@ -0,0 +1,33 @@
|
|||
\begin{juliacode}
|
||||
using Plots
|
||||
gr()
|
||||
x = linspace(0, 2*pi)
|
||||
println(x)
|
||||
\end{juliacode}
|
||||
\begin{juliaout}
|
||||
0.0:0.1282282715750936:6.283185307179586
|
||||
\end{juliaout}
|
||||
|
||||
\begin{juliacode}
|
||||
p = plot(x, sin.(x), size =(900,300))
|
||||
p
|
||||
\end{juliacode}
|
||||
\includegraphics[width=\linewidth]{figures/plotsjl_test_gr_1_1.pdf}
|
||||
|
||||
\begin{juliacode}
|
||||
plot(x, sin.(x))
|
||||
\end{juliacode}
|
||||
\includegraphics[width=\linewidth]{figures/plotsjl_test_gr_2_1.pdf}
|
||||
|
||||
\begin{juliacode}
|
||||
plot(rand(100) / 3,reg=true,fill=(0,:green))
|
||||
scatter!(rand(100),markersize=6,c=:orange)
|
||||
\end{juliacode}
|
||||
\includegraphics[width=\linewidth]{figures/plotsjl_test_gr_3_1.pdf}
|
||||
|
||||
\begin{figure}[htpb]
|
||||
\center
|
||||
\includegraphics[width=\linewidth]{figures/plotsjl_test_gr_random_1.pdf}
|
||||
\caption{A random walk.}
|
||||
\label{fig:random}
|
||||
\end{figure}
|
||||
|
|
@ -5,12 +5,13 @@ using Plots
|
|||
gr()
|
||||
x = linspace(0, 2*pi)
|
||||
println(x)
|
||||
p = plot(x, sin(x), size =(900,300))
|
||||
p = plot(x, sin.(x), size =(900,300))
|
||||
p
|
||||
```
|
||||
|
||||
|
||||
```julia; term=true
|
||||
plot(x, sin(x))
|
||||
```julia
|
||||
plot(x, sin.(x))
|
||||
```
|
||||
|
||||
```julia
|
||||
|
|
@ -18,11 +19,6 @@ plot(rand(100) / 3,reg=true,fill=(0,:green))
|
|||
scatter!(rand(100),markersize=6,c=:orange)
|
||||
```
|
||||
|
||||
```julia; term=true
|
||||
plot(rand(100) / 3,reg=true,fill=(0,:green))
|
||||
scatter!(rand(100),markersize=6,c=:orange)
|
||||
```
|
||||
|
||||
|
||||
```{julia;echo=false; fig_cap="A random walk."; label="random"}
|
||||
plot(cumsum(randn(1000, 1)))
|
||||
|
|
|
|||
|
|
@ -1,6 +1,5 @@
|
|||
using Weave
|
||||
using Base.Test
|
||||
import Plots
|
||||
|
||||
function pljtest(source, resfile, doctype)
|
||||
weave("documents/$source", out_path = "documents/plotsjl/$resfile", doctype=doctype)
|
||||
|
|
@ -10,7 +9,8 @@ function pljtest(source, resfile, doctype)
|
|||
rm("documents/plotsjl/$resfile")
|
||||
end
|
||||
|
||||
pljtest("plotsjl_test.jmd", "plotsjl_test.md", "pandoc")
|
||||
pljtest("plotsjl_test.jmd", "plotsjl_test.tex", "tex")
|
||||
#pljtest("plotsjl_test.jmd", "plotsjl_test.md", "pandoc")
|
||||
#pljtest("plotsjl_test.jmd", "plotsjl_test.tex", "tex")
|
||||
|
||||
pljtest("plotsjl_test_gr.jmd", "plotsjl_test_gr.md", "pandoc")
|
||||
pljtest("plotsjl_test_gr.jmd", "plotsjl_test_gr.tex", "tex")
|
||||
|
|
@ -1,4 +1,3 @@
|
|||
import PyPlot
|
||||
using Weave
|
||||
using Base.Test
|
||||
|
||||
|
|
@ -14,20 +13,20 @@ include("formatter_test.jl")
|
|||
info("Testing rich output")
|
||||
include("rich_output.jl")
|
||||
|
||||
if VERSION < v"0.6-"
|
||||
info("Test: Caching")
|
||||
include("cache_test.jl")
|
||||
|
||||
info("Test: Chunk options with Gadfly")
|
||||
include("chunk_opts_gadfly.jl")
|
||||
#info("Test: Caching")
|
||||
#include("cache_test.jl")
|
||||
|
||||
info("Test: Weaving with Gadfly")
|
||||
include("gadfly_formats.jl")
|
||||
#info("Test: Chunk options with Gadfly")
|
||||
#include("chunk_opts_gadfly.jl")
|
||||
|
||||
info("Test: Weaving with PyPlot")
|
||||
include("pyplot_formats.jl")
|
||||
#info("Test: Weaving with Gadfly")
|
||||
#include("gadfly_formats.jl")
|
||||
|
||||
#info("Test: Weaving with PyPlot")
|
||||
#include("pyplot_formats.jl")
|
||||
|
||||
info("Test: Weaving with Plots.jl")
|
||||
include("plotsjl_test.jl")
|
||||
include("publish_test.jl")
|
||||
|
||||
info("Test: Weaving with Plots.jl")
|
||||
include("plotsjl_test.jl")
|
||||
include("publish_test.jl")
|
||||
end
|
||||
|
|
|
|||
Loading…
Reference in New Issue