mirror of https://github.com/mpastell/Weave.jl
Fix Gadfly and Plots, use Compat.invokelatest
parent
562968f9be
commit
5c83309aa3
|
|
@ -27,7 +27,9 @@ and figures.
|
|||
**Citing Weave:** *Pastell, Matti. 2017. Weave.jl: Scientific Reports Using Julia. The Journal of Open Source Software. http://dx.doi.org/10.21105/joss.00204*
|
||||
|
||||
|
||||
**Note about Julia 0.6**: Weave has been updated to support 0.6, but most of the depencies and plotting libraries have not. Plotting currently (14th March 2017) only works using PyPlot master.
|
||||
**Note about Julia 0.6**: Weave has been updated to support 0.6, current master
|
||||
requires using `Compat` master (`Compat.invokelatest`). PyPlot, Gadfly and Plots now
|
||||
also work (12th May 2017).
|
||||
|
||||
|
||||
|
||||
|
|
|
|||
|
|
@ -1,6 +1,7 @@
|
|||
__precompile__(false)
|
||||
module Weave
|
||||
import Highlights
|
||||
using Compat
|
||||
|
||||
"""
|
||||
`list_out_formats()`
|
||||
|
|
|
|||
|
|
@ -1,3 +1,5 @@
|
|||
using Compat
|
||||
|
||||
#Contains report global properties
|
||||
type Report <: Display
|
||||
cwd::AbstractString
|
||||
|
|
@ -31,13 +33,7 @@ function Base.display(report::Report, data)
|
|||
for m in report.mimetypes
|
||||
if mimewritable(m, data)
|
||||
try
|
||||
if VERSION >= v"0.6.0-dev.1671"
|
||||
new_dp(x, y, z) = eval(
|
||||
Expr(:call, (x, y, z) -> display(x, y, z), x, y, z))
|
||||
new_dp(report, m, data)
|
||||
else
|
||||
display(report, m, data)
|
||||
end
|
||||
@compat Compat.invokelatest(display, report, m, data)
|
||||
catch e
|
||||
warn("Failed to display data in \"$m\" format")
|
||||
continue
|
||||
|
|
|
|||
|
|
@ -1,4 +1,5 @@
|
|||
import Mustache, Highlights, Documenter
|
||||
using Compat
|
||||
|
||||
function format(doc::WeaveDoc)
|
||||
formatted = AbstractString[]
|
||||
|
|
@ -128,7 +129,9 @@ end
|
|||
|
||||
function format_chunk(chunk::DocChunk, formatdict, docformat::JMarkdown2HTML)
|
||||
text = format_chunk(chunk, formatdict, nothing)
|
||||
m = Base.Markdown.parse(text)
|
||||
#invokelatest seems to be needed here
|
||||
#to fix "invalid age range" on 0.6 #21653
|
||||
m = @compat Compat.invokelatest(Base.Markdown.parse, text)
|
||||
return string(Documenter.Writers.HTMLWriter.mdconvert(m))
|
||||
end
|
||||
|
||||
|
|
|
|||
17
src/run.jl
17
src/run.jl
|
|
@ -212,13 +212,7 @@ function run_code(chunk::CodeChunk, report::Report, SandBox::Module)
|
|||
|
||||
#Save figures only in the end of chunk for PyPlot
|
||||
if rcParams[:plotlib] == "PyPlot"
|
||||
#Fix "world-age" issue
|
||||
if VERSION >= v"0.6-alpha"
|
||||
savep(x) = eval(Expr(:call, x-> savefigs_pyplot(x), x))
|
||||
savep(report)
|
||||
else
|
||||
savefigs_pyplot(report)
|
||||
end
|
||||
@compat Compat.invokelatest(savefigs_pyplot, report)
|
||||
end
|
||||
|
||||
return results
|
||||
|
|
@ -276,8 +270,7 @@ end
|
|||
|
||||
|
||||
function eval_chunk(chunk::CodeChunk, report::Report, SandBox::Module)
|
||||
|
||||
|
||||
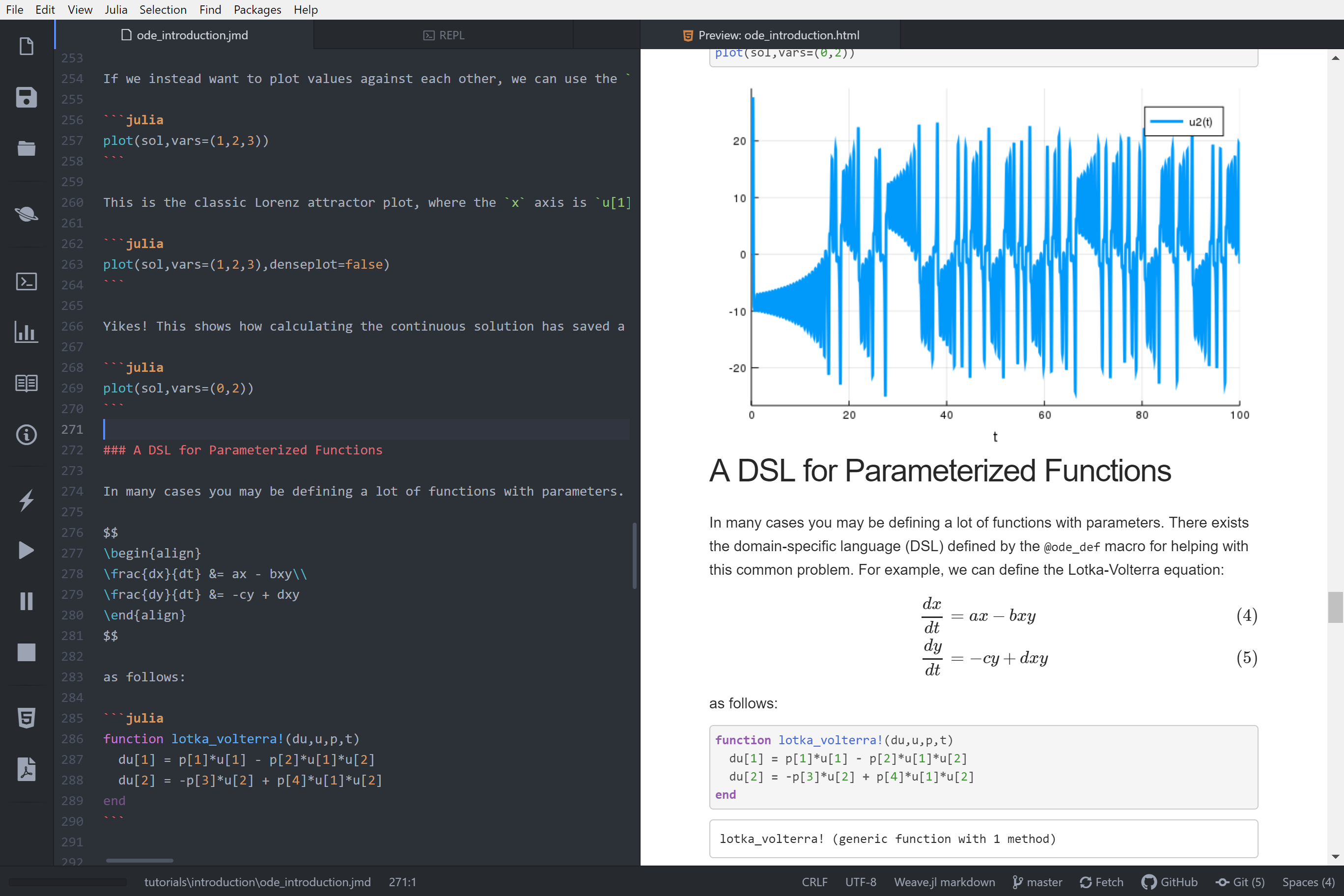
|
||||
if !chunk.options[:eval]
|
||||
chunk.output = ""
|
||||
chunk.options[:fig] = false
|
||||
|
|
@ -286,7 +279,7 @@ function eval_chunk(chunk::CodeChunk, report::Report, SandBox::Module)
|
|||
|
||||
#Run preexecute_hooks
|
||||
for hook in preexecute_hooks
|
||||
chunk = hook(chunk)
|
||||
chunk = @compat Compat.invokelatest(hook, chunk)
|
||||
end
|
||||
|
||||
report.fignum = 1
|
||||
|
|
@ -301,7 +294,7 @@ function eval_chunk(chunk::CodeChunk, report::Report, SandBox::Module)
|
|||
|
||||
#Run post_execute chunks
|
||||
for hook in postexecute_hooks
|
||||
chunk = hook(chunk)
|
||||
chunk = @compat Compat.invokelatest(hook, chunk)
|
||||
end
|
||||
|
||||
if chunk.options[:term]
|
||||
|
|
@ -492,7 +485,7 @@ function detect_plotlib(chunk::CodeChunk)
|
|||
if isdefined(:Plots)
|
||||
init_plotting("Plots")
|
||||
#Need to set size before plots are created
|
||||
plots_set_size(chunk)
|
||||
@compat Compat.invokelatest(plots_set_size, chunk)
|
||||
return
|
||||
end
|
||||
|
||||
|
|
|
|||
|
|
@ -27,8 +27,8 @@ if VERSION < v"0.6-"
|
|||
|
||||
info("Test: Weaving with PyPlot")
|
||||
include("pyplot_formats.jl")
|
||||
|
||||
info("Test: Weaving with Plots.jl")
|
||||
include("plotsjl_test.jl")
|
||||
include("publish_test.jl")
|
||||
end
|
||||
|
||||
info("Test: Weaving with Plots.jl")
|
||||
include("plotsjl_test.jl")
|
||||
include("publish_test.jl")
|
||||
|
|
|
|||
Loading…
Reference in New Issue