block style of highligh.js.
+ * We have to override the padding and the background-color, since we style this
+ * part ourselves. Specifically, we style the surrounding the , while
+ * highlight.js applies the .hljs style directly to the tag.
+ */
+.hljs {
+ background-color: transparent;
+ padding: 0;
+}
+
+@media only screen and (max-width: 768px) {
+ nav.toc {
+ position: fixed;
+ width: 16em;
+ left: -16em;
+ -webkit-overflow-scrolling: touch;
+ -webkit-transition-property: left; /* Safari */
+ -webkit-transition-duration: 0.3s; /* Safari */
+ transition-property: left;
+ transition-duration: 0.3s;
+ -webkit-transition-timing-function: ease-out; /* Safari */
+ transition-timing-function: ease-out;
+ z-index: 2;
+ box-shadow: 5px 0px 5px 0px rgb(210,210,210);
+ }
+
+ nav.toc.show {
+ left: 0;
+ }
+
+ article {
+ margin-left: 0;
+ padding: 3em 0.9em 0 0.9em; /* top right bottom left */
+ overflow-wrap: break-word;
+ }
+
+ article > header {
+ position: fixed;
+ left: 0;
+ z-index: 1;
+ }
+
+ article > header nav, hr {
+ display: none;
+ }
+
+ article > header div#topbar {
+ display: block; /* is mobile */
+ position: fixed;
+ width: 100%;
+ height: 1.5em;
+ padding-top: 1em;
+ padding-bottom: 1em;
+ background-color: #fcfcfc;
+ box-shadow: 0 1px 3px rgba(0,0,0,.26);
+ top: 0;
+ -webkit-transition-property: top; /* Safari */
+ -webkit-transition-duration: 0.3s; /* Safari */
+ transition-property: top;
+ transition-duration: 0.3s;
+ }
+
+ article > header div#topbar.headroom--unpinned.headroom--not-top.headroom--not-bottom {
+ top: -4em;
+ -webkit-transition-property: top; /* Safari */
+ -webkit-transition-duration: 0.7s; /* Safari */
+ transition-property: top;
+ transition-duration: 0.7s;
+ }
+
+ article > header div#topbar span {
+ width: 80%;
+ height: 1.5em;
+ margin-top: -0.1em;
+ margin-left: 0.9em;
+ font-size: 1.2em;
+ overflow: hidden;
+ }
+
+ article > header div#topbar a.fa-bars {
+ float: right;
+ padding: 0.6em;
+ margin-top: -0.6em;
+ margin-right: 0.3em;
+ font-size: 1.5em;
+ }
+
+ article > header div#topbar a.fa-bars:visited {
+ color: #3091d1;
+ }
+
+ article table {
+ overflow-x: auto;
+ display: block;
+ }
+
+ article div.MathJax_Display {
+ overflow: scroll;
+ }
+
+ article span.MathJax {
+ overflow: hidden;
+ }
+}
+
+@media only screen and (max-width: 320px) {
+ body {
+ font-size: 15px;
+ }
+}
diff --git a/v0.8.1/assets/documenter.js b/v0.8.1/assets/documenter.js
new file mode 100644
index 0000000..761ae29
--- /dev/null
+++ b/v0.8.1/assets/documenter.js
@@ -0,0 +1,132 @@
+/*
+ * Part of Documenter.jl
+ * https://github.com/JuliaDocs/Documenter.jl
+ *
+ * License: MIT
+ */
+
+requirejs.config({
+ paths: {
+ 'jquery': 'https://cdnjs.cloudflare.com/ajax/libs/jquery/3.1.1/jquery.min',
+ 'jqueryui': 'https://cdnjs.cloudflare.com/ajax/libs/jqueryui/1.12.0/jquery-ui.min',
+ 'headroom': 'https://cdnjs.cloudflare.com/ajax/libs/headroom/0.9.3/headroom.min',
+ 'mathjax': 'https://cdnjs.cloudflare.com/ajax/libs/mathjax/2.7.1/MathJax.js?config=TeX-AMS_HTML',
+ 'highlight': 'https://cdnjs.cloudflare.com/ajax/libs/highlight.js/9.12.0/highlight.min',
+ 'highlight-julia': 'https://cdnjs.cloudflare.com/ajax/libs/highlight.js/9.12.0/languages/julia.min',
+ 'highlight-julia-repl': 'https://cdnjs.cloudflare.com/ajax/libs/highlight.js/9.12.0/languages/julia-repl.min',
+ },
+ shim: {
+ 'mathjax' : {
+ exports: "MathJax"
+ },
+ 'highlight-julia': ['highlight'],
+ 'highlight-julia-repl': ['highlight'],
+ }
+});
+
+// Load MathJax
+require(['mathjax'], function(MathJax) {
+ MathJax.Hub.Config({
+ "tex2jax": {
+ inlineMath: [['$','$'], ['\\(','\\)']],
+ processEscapes: true
+ }
+ });
+ MathJax.Hub.Config({
+ config: ["MMLorHTML.js"],
+ jax: [
+ "input/TeX",
+ "output/HTML-CSS",
+ "output/NativeMML"
+ ],
+ extensions: [
+ "MathMenu.js",
+ "MathZoom.js",
+ "TeX/AMSmath.js",
+ "TeX/AMSsymbols.js",
+ "TeX/autobold.js",
+ "TeX/autoload-all.js"
+ ]
+ });
+ MathJax.Hub.Config({
+ TeX: { equationNumbers: { autoNumber: "AMS" } }
+ });
+})
+
+require(['jquery', 'highlight', 'highlight-julia', 'highlight-julia-repl'], function($, hljs) {
+ $(document).ready(function() {
+ hljs.initHighlighting();
+ })
+
+})
+
+// update the version selector with info from the siteinfo.js and ../versions.js files
+require(['jquery'], function($) {
+ $(document).ready(function() {
+ var version_selector = $("#version-selector");
+
+ // add the current version to the selector based on siteinfo.js, but only if the selector is empty
+ if (typeof DOCUMENTER_CURRENT_VERSION !== 'undefined' && $('#version-selector > option').length == 0) {
+ var option = $("");
+ version_selector.append(option);
+ }
+
+ if (typeof DOC_VERSIONS !== 'undefined') {
+ var existing_versions = $('#version-selector > option');
+ var existing_versions_texts = existing_versions.map(function(i,x){return x.text});
+ DOC_VERSIONS.forEach(function(each) {
+ var version_url = documenterBaseURL + "/../" + each;
+ var existing_id = $.inArray(each, existing_versions_texts);
+ // if not already in the version selector, add it as a new option,
+ // otherwise update the old option with the URL and enable it
+ if (existing_id == -1) {
+ var option = $("");
+ version_selector.append(option);
+ } else {
+ var option = existing_versions[existing_id];
+ option.value = version_url;
+ option.disabled = false;
+ }
+ });
+ }
+
+ // only show the version selector if the selector has been populated
+ if ($('#version-selector > option').length > 0) {
+ version_selector.css("visibility", "visible");
+ }
+
+ // Scroll the navigation bar to the currently selected menu item
+ $("nav.toc > ul").get(0).scrollTop = $(".current").get(0).offsetTop - $("nav.toc > ul").get(0).offsetTop;
+ })
+
+})
+
+// mobile
+require(['jquery', 'headroom'], function($, Headroom) {
+ $(document).ready(function() {
+ var navtoc = $("nav.toc");
+ $("nav.toc li.current a.toctext").click(function() {
+ navtoc.toggleClass('show');
+ });
+ $("article > header div#topbar a.fa-bars").click(function(ev) {
+ ev.preventDefault();
+ navtoc.toggleClass('show');
+ if (navtoc.hasClass('show')) {
+ var title = $("article > header div#topbar span").text();
+ $("nav.toc ul li a:contains('" + title + "')").focus();
+ }
+ });
+ $("article#docs").bind('click', function(ev) {
+ if ($(ev.target).is('div#topbar a.fa-bars')) {
+ return;
+ }
+ if (navtoc.hasClass('show')) {
+ navtoc.removeClass('show');
+ }
+ });
+ if ($("article > header div#topbar").css('display') == 'block') {
+ var headroom = new Headroom(document.querySelector("article > header div#topbar"), {"tolerance": {"up": 10, "down": 10}});
+ headroom.init();
+ }
+ })
+})
diff --git a/v0.8.1/assets/search.js b/v0.8.1/assets/search.js
new file mode 100644
index 0000000..f99b378
--- /dev/null
+++ b/v0.8.1/assets/search.js
@@ -0,0 +1,250 @@
+/*
+ * Part of Documenter.jl
+ * https://github.com/JuliaDocs/Documenter.jl
+ *
+ * License: MIT
+ */
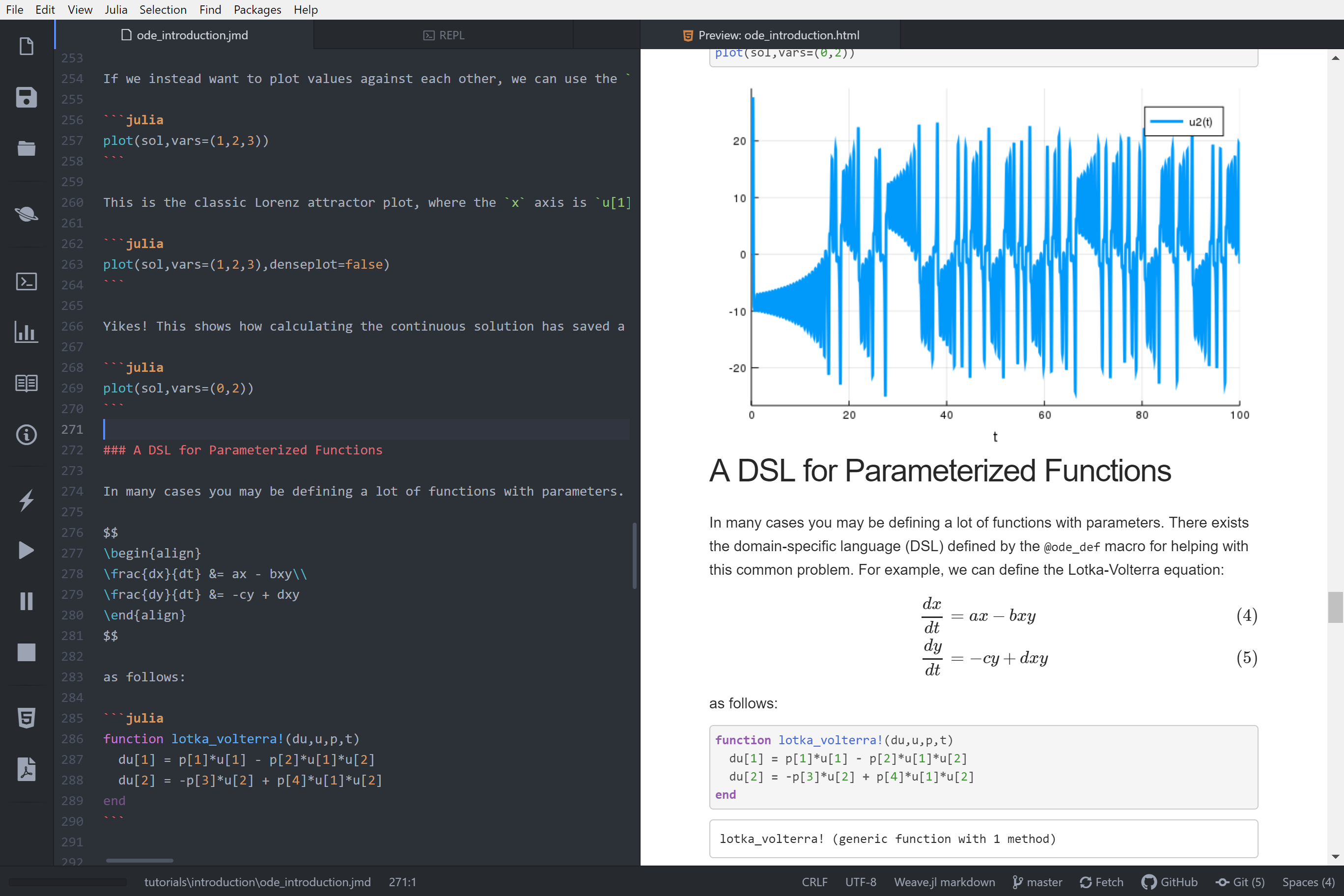
+
+// parseUri 1.2.2
+// (c) Steven Levithan
+// MIT License
+function parseUri (str) {
+ var o = parseUri.options,
+ m = o.parser[o.strictMode ? "strict" : "loose"].exec(str),
+ uri = {},
+ i = 14;
+
+ while (i--) uri[o.key[i]] = m[i] || "";
+
+ uri[o.q.name] = {};
+ uri[o.key[12]].replace(o.q.parser, function ($0, $1, $2) {
+ if ($1) uri[o.q.name][$1] = $2;
+ });
+
+ return uri;
+};
+parseUri.options = {
+ strictMode: false,
+ key: ["source","protocol","authority","userInfo","user","password","host","port","relative","path","directory","file","query","anchor"],
+ q: {
+ name: "queryKey",
+ parser: /(?:^|&)([^&=]*)=?([^&]*)/g
+ },
+ parser: {
+ strict: /^(?:([^:\/?#]+):)?(?:\/\/((?:(([^:@]*)(?::([^:@]*))?)?@)?([^:\/?#]*)(?::(\d*))?))?((((?:[^?#\/]*\/)*)([^?#]*))(?:\?([^#]*))?(?:#(.*))?)/,
+ loose: /^(?:(?![^:@]+:[^:@\/]*@)([^:\/?#.]+):)?(?:\/\/)?((?:(([^:@]*)(?::([^:@]*))?)?@)?([^:\/?#]*)(?::(\d*))?)(((\/(?:[^?#](?![^?#\/]*\.[^?#\/.]+(?:[?#]|$)))*\/?)?([^?#\/]*))(?:\?([^#]*))?(?:#(.*))?)/
+ }
+};
+
+requirejs.config({
+ paths: {
+ 'jquery': 'https://cdnjs.cloudflare.com/ajax/libs/jquery/3.1.1/jquery.min',
+ 'lunr': 'https://cdnjs.cloudflare.com/ajax/libs/lunr.js/2.3.1/lunr.min',
+ 'lodash': 'https://cdnjs.cloudflare.com/ajax/libs/lodash.js/4.17.4/lodash.min',
+ }
+});
+
+var currentScript = document.currentScript;
+
+require(["jquery", "lunr", "lodash"], function($, lunr, _) {
+ $("#search-form").submit(function(e) {
+ e.preventDefault()
+ })
+
+ // list below is the lunr 2.1.3 list minus the intersect with names(Base)
+ // (all, any, get, in, is, which) and (do, else, for, let, where, while, with)
+ // ideally we'd just filter the original list but it's not available as a variable
+ lunr.stopWordFilter = lunr.generateStopWordFilter([
+ 'a',
+ 'able',
+ 'about',
+ 'across',
+ 'after',
+ 'almost',
+ 'also',
+ 'am',
+ 'among',
+ 'an',
+ 'and',
+ 'are',
+ 'as',
+ 'at',
+ 'be',
+ 'because',
+ 'been',
+ 'but',
+ 'by',
+ 'can',
+ 'cannot',
+ 'could',
+ 'dear',
+ 'did',
+ 'does',
+ 'either',
+ 'ever',
+ 'every',
+ 'from',
+ 'got',
+ 'had',
+ 'has',
+ 'have',
+ 'he',
+ 'her',
+ 'hers',
+ 'him',
+ 'his',
+ 'how',
+ 'however',
+ 'i',
+ 'if',
+ 'into',
+ 'it',
+ 'its',
+ 'just',
+ 'least',
+ 'like',
+ 'likely',
+ 'may',
+ 'me',
+ 'might',
+ 'most',
+ 'must',
+ 'my',
+ 'neither',
+ 'no',
+ 'nor',
+ 'not',
+ 'of',
+ 'off',
+ 'often',
+ 'on',
+ 'only',
+ 'or',
+ 'other',
+ 'our',
+ 'own',
+ 'rather',
+ 'said',
+ 'say',
+ 'says',
+ 'she',
+ 'should',
+ 'since',
+ 'so',
+ 'some',
+ 'than',
+ 'that',
+ 'the',
+ 'their',
+ 'them',
+ 'then',
+ 'there',
+ 'these',
+ 'they',
+ 'this',
+ 'tis',
+ 'to',
+ 'too',
+ 'twas',
+ 'us',
+ 'wants',
+ 'was',
+ 'we',
+ 'were',
+ 'what',
+ 'when',
+ 'who',
+ 'whom',
+ 'why',
+ 'will',
+ 'would',
+ 'yet',
+ 'you',
+ 'your'
+ ])
+
+ // add . as a separator, because otherwise "title": "Documenter.Anchors.add!"
+ // would not find anything if searching for "add!", only for the entire qualification
+ lunr.tokenizer.separator = /[\s\-\.]+/
+
+ // custom trimmer that doesn't strip @ and !, which are used in julia macro and function names
+ lunr.trimmer = function (token) {
+ return token.update(function (s) {
+ return s.replace(/^[^a-zA-Z0-9@!]+/, '').replace(/[^a-zA-Z0-9@!]+$/, '')
+ })
+ }
+
+ lunr.Pipeline.registerFunction(lunr.stopWordFilter, 'juliaStopWordFilter')
+ lunr.Pipeline.registerFunction(lunr.trimmer, 'juliaTrimmer')
+
+ var index = lunr(function () {
+ this.ref('location')
+ this.field('title')
+ this.field('text')
+ documenterSearchIndex['docs'].forEach(function(e) {
+ this.add(e)
+ }, this)
+ })
+ var store = {}
+
+ documenterSearchIndex['docs'].forEach(function(e) {
+ store[e.location] = {title: e.title, category: e.category}
+ })
+
+ $(function(){
+ function update_search(querystring) {
+ tokens = lunr.tokenizer(querystring)
+ results = index.query(function (q) {
+ tokens.forEach(function (t) {
+ q.term(t.toString(), {
+ fields: ["title"],
+ boost: 100,
+ usePipeline: false,
+ editDistance: 0,
+ wildcard: lunr.Query.wildcard.NONE
+ })
+ q.term(t.toString(), {
+ fields: ["title"],
+ boost: 10,
+ usePipeline: false,
+ editDistance: 2,
+ wildcard: lunr.Query.wildcard.NONE
+ })
+ q.term(t.toString(), {
+ fields: ["text"],
+ boost: 1,
+ usePipeline: true,
+ editDistance: 0,
+ wildcard: lunr.Query.wildcard.NONE
+ })
+ })
+ })
+ $('#search-info').text("Number of results: " + results.length)
+ $('#search-results').empty()
+ results.forEach(function(result) {
+ data = store[result.ref]
+ link = $('')
+ link.text(data.title)
+ link.attr('href', documenterBaseURL+'/'+result.ref)
+ cat = $('('+data.category+')')
+ li = $('').append(link).append(" ").append(cat)
+ $('#search-results').append(li)
+ })
+ }
+
+ function update_search_box() {
+ querystring = $('#search-query').val()
+ update_search(querystring)
+ }
+
+ $('#search-query').keyup(_.debounce(update_search_box, 250))
+ $('#search-query').change(update_search_box)
+
+ search_query_uri = parseUri(window.location).queryKey["q"]
+ if(search_query_uri !== undefined) {
+ search_query = decodeURIComponent(search_query_uri.replace(/\+/g, '%20'))
+ $("#search-query").val(search_query)
+ }
+ update_search_box();
+ })
+})
diff --git a/v0.8.1/chunk_options/index.html b/v0.8.1/chunk_options/index.html
new file mode 100644
index 0000000..e65b999
--- /dev/null
+++ b/v0.8.1/chunk_options/index.html
@@ -0,0 +1,7 @@
+
+Chunk options · Weave.jl
Chunk options
I've mostly followed Knitr's naming for chunk options, but not all options are implemented.
Options are separated using ";" and need to be valid Julia expressions. Example: markdown code chunk that saves and displays a 12 cm wide image and hides the source code:
julia; out_width="12cm"; echo=false
Weave currently supports the following chunk options with the following defaults:
Options for code
echo = true. Echo the code in the output document. If false the source code will be hidden.results = "markup". The output format of the printed results. "markup" for literal block, "hidden" for hidden results or anything else for raw output (I tend to use ‘tex’ for Latex and ‘rst’ for rest. Raw output is useful if you wan’t to e.g. create tables from code chunks.eval = true. Evaluate the code chunk. If false the chunk won’t be executed.term=false. If true the output emulates a REPL session. Otherwise only stdout and figures will be included in output.label. Chunk label, will be used for figure labels in Latex as fig:labelwrap = true. Wrap long lines from output.line_width = 75. Line width for wrapped lines.cache = false. Cache results, depends on cache parameter on weave function.hold = false. Hold all results until the end of the chunk.tangle = true. Set tangle to false to exclude chunk from tangled code.
Options for figures
fig_width. Figure width passed to plotting library e.g. 800fig_height Figure height passed to plotting libraryout_width. Width of saved figure in output markup e.g. "50%", "12cm", \\0.5linewidthout_height. Height of saved figure in output markupdpi=96. Resolution of saved figures.fig_cap. Figure caption.label. Chunk label, will be used for figure labels in Latex as fig:labelfig_ext. File extension (format) of saved figures.fig_pos="htpb". Figure position in Latex. fig_env="figure". Figure environment in Latex.
Set default chunk options
You can set the default chunk options (and weave arguments) for a document using the YAML header options field. e.g to set the default out_width of all figures you can use:
---
+options:
+ out_width : 50%
+---
You can also set or change the default chunk options for a document either before weave using the set_chunk_defaults function.
set_chunk_defaults(opts)
+get_chunk_defaults()
+restore_chunk_defaults()
FIR filter design with Julia
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ FIR filter design with Julia
+ Matti Pastell
+ 21th April 2016
+
+
+ Introduction
+This an example of a julia script that can be published using Weave. The script can be executed normally using Julia or published to HTML or pdf with Weave. Text is written in markdown in lines starting with "#' " and code is executed and results are included in the published document.
+Notice that you don't need to define chunk options, but you can using #+. just before code e.g. #+ term=True, caption='Fancy plots.'. If you're viewing the published version have a look at the source to see the markup.
+FIR Filter Design
+We'll implement lowpass, highpass and ' bandpass FIR filters. If you want to read more about DSP I highly recommend The Scientist and Engineer's Guide to Digital Signal Processing which is freely available online.
+Calculating frequency response
+DSP.jl package doesn't (yet) have a method to calculate the the frequency response of a FIR filter so we define it:
+
+
+
+using Plots, DSP
+gr()
+
+function FIRfreqz(b::Array, w = range(0, stop=π, length=1024))
+ n = length(w)
+ h = Array{ComplexF32}(undef, n)
+ sw = 0
+ for i = 1:n
+ for j = 1:length(b)
+ sw += b[j]*exp(-im*w[i])^-j
+ end
+ h[i] = sw
+ sw = 0
+ end
+ return h
+end
+
+
+
+
+FIRfreqz (generic function with 2 methods)
+
+
+
+Design Lowpass FIR filter
+Designing a lowpass FIR filter is very simple to do with DSP.jl, all you need to do is to define the window length, cut off frequency and the window. We will define a lowpass filter with cut off frequency at 5Hz for a signal sampled at 20 Hz. We will use the Hamming window, which is defined as: $w(n) = \alpha - \beta\cos\frac{2\pi n}{N-1}$, where $\alpha=0.54$ and $\beta=0.46$
+
+
+
+fs = 20
+f = digitalfilter(Lowpass(5, fs = fs), FIRWindow(hamming(61)))
+w = range(0, stop=pi, length=1024)
+h = FIRfreqz(f, w)
+
+
+
+
+1024-element Array{Complex{Float32},1}:
+ 1.0f0 + 0.0f0im
+ 0.99546844f0 + 0.095055714f0im
+ 0.98191506f0 + 0.1892486f0im
+ 0.95946306f0 + 0.28172377f0im
+ 0.9283168f0 + 0.37164196f0im
+ 0.8887594f0 + 0.45818728f0im
+ 0.84115064f0 + 0.54057467f0im
+ 0.7859234f0 + 0.618057f0im
+ 0.72357976f0 + 0.6899319f0im
+ 0.65468615f0 + 0.7555481f0im
+ ⋮
+ 0.00043952762f0 - 0.00041908873f0im
+ 0.0005152718f0 - 0.00040521423f0im
+ 0.0005873293f0 - 0.00037745363f0im
+ 0.0006531789f0 - 0.0003367371f0im
+ 0.0007105166f0 - 0.00028444792f0im
+ 0.0007573364f0 - 0.00022237403f0im
+ 0.0007920005f0 - 0.00015264557f0im
+ 0.0008132961f0 - 7.766036f-5im
+ 0.0008204784f0 - 3.1148685f-18im
+
+
+
+Plot the frequency and impulse response
+The next code chunk is executed in term mode, see the script for syntax.
+
+
+julia> h_db = log10.(abs.(h));
+
+julia> ws = w/pi*(fs/2)
+0.0:0.009775171065493646:10.0
+
+
+
+
+plot(ws, h_db,
+ xlabel = "Frequency (Hz)", ylabel = "Magnitude (db)")
+
+
+
+ +
+
+
+And again with default options
+
+
+
+h_phase = unwrap(-atan.(imag.(h),real.(h)))
+plot(ws, h_phase,
+ xlabel = "Frequency (Hz)", ylabel = "Phase (radians)")
+
+
+
+ +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
diff --git a/v0.8.1/examples/FIR_design.jmd b/v0.8.1/examples/FIR_design.jmd
new file mode 100644
index 0000000..2266480
--- /dev/null
+++ b/v0.8.1/examples/FIR_design.jmd
@@ -0,0 +1,99 @@
+---
+title: FIR filter design with Julia
+author: Matti Pastell
+date: 21th April 2016
+---
+
+
+# Introduction
+
+
+This an example of a julia script that can be published using
+[Weave](http://mpastell.github.io/Weave.jl/latest/usage/).
+The script can be executed normally using Julia
+or published to HTML or pdf with Weave.
+Text is written in markdown in lines starting with "`#'` " and code
+is executed and results are included in the published document.
+
+
+Notice that you don't need to define chunk options, but you can using
+`#+`. just before code e.g. `#+ term=True, caption='Fancy plots.'`.
+If you're viewing the published version have a look at the
+[source](FIR_design_plots.jl) to see the markup.
+
+
+
+# FIR Filter Design
+
+
+We'll implement lowpass, highpass and ' bandpass FIR filters. If
+you want to read more about DSP I highly recommend [The Scientist
+and Engineer's Guide to Digital Signal
+Processing](http://www.dspguide.com/) which is freely available
+online.
+
+
+## Calculating frequency response
+
+
+DSP.jl package doesn't (yet) have a method to calculate the
+the frequency response of a FIR filter so we define it:
+
+```julia
+using Plots, DSP
+gr()
+
+function FIRfreqz(b::Array, w = range(0, stop=π, length=1024))
+ n = length(w)
+ h = Array{ComplexF32}(undef, n)
+ sw = 0
+ for i = 1:n
+ for j = 1:length(b)
+ sw += b[j]*exp(-im*w[i])^-j
+ end
+ h[i] = sw
+ sw = 0
+ end
+ return h
+end
+```
+
+## Design Lowpass FIR filter
+
+
+Designing a lowpass FIR filter is very simple to do with DSP.jl, all you
+need to do is to define the window length, cut off frequency and the
+window. We will define a lowpass filter with cut off frequency at 5Hz for a signal
+sampled at 20 Hz.
+We will use the Hamming window, which is defined as:
+$w(n) = \alpha - \beta\cos\frac{2\pi n}{N-1}$, where $\alpha=0.54$ and $\beta=0.46$
+
+```julia
+fs = 20
+f = digitalfilter(Lowpass(5, fs = fs), FIRWindow(hamming(61)))
+w = range(0, stop=pi, length=1024)
+h = FIRfreqz(f, w)
+```
+
+## Plot the frequency and impulse response
+
+
+The next code chunk is executed in term mode, see the [script](FIR_design.jl) for syntax.
+
+```julia; term=true
+h_db = log10.(abs.(h));
+ws = w/pi*(fs/2)
+```
+
+```julia
+plot(ws, h_db,
+ xlabel = "Frequency (Hz)", ylabel = "Magnitude (db)")
+```
+
+And again with default options
+
+```julia
+h_phase = unwrap(-atan.(imag.(h),real.(h)))
+plot(ws, h_phase,
+ xlabel = "Frequency (Hz)", ylabel = "Phase (radians)")
+```
diff --git a/v0.8.1/examples/FIR_design.md b/v0.8.1/examples/FIR_design.md
new file mode 100644
index 0000000..d9cae50
--- /dev/null
+++ b/v0.8.1/examples/FIR_design.md
@@ -0,0 +1,152 @@
+---
+title: FIR filter design with Julia
+author: Matti Pastell
+date: 21th April 2016
+---
+
+
+# Introduction
+
+
+This an example of a julia script that can be published using
+[Weave](http://mpastell.github.io/Weave.jl/latest/usage/).
+The script can be executed normally using Julia
+or published to HTML or pdf with Weave.
+Text is written in markdown in lines starting with "`#'` " and code
+is executed and results are included in the published document.
+
+
+Notice that you don't need to define chunk options, but you can using
+`#+`. just before code e.g. `#+ term=True, caption='Fancy plots.'`.
+If you're viewing the published version have a look at the
+[source](FIR_design_plots.jl) to see the markup.
+
+
+
+# FIR Filter Design
+
+
+We'll implement lowpass, highpass and ' bandpass FIR filters. If
+you want to read more about DSP I highly recommend [The Scientist
+and Engineer's Guide to Digital Signal
+Processing](http://www.dspguide.com/) which is freely available
+online.
+
+
+## Calculating frequency response
+
+
+DSP.jl package doesn't (yet) have a method to calculate the
+the frequency response of a FIR filter so we define it:
+
+~~~~{.julia}
+using Plots, DSP
+gr()
+
+function FIRfreqz(b::Array, w = range(0, stop=π, length=1024))
+ n = length(w)
+ h = Array{ComplexF32}(undef, n)
+ sw = 0
+ for i = 1:n
+ for j = 1:length(b)
+ sw += b[j]*exp(-im*w[i])^-j
+ end
+ h[i] = sw
+ sw = 0
+ end
+ return h
+end
+~~~~~~~~~~~~~
+
+
+~~~~
+FIRfreqz (generic function with 2 methods)
+~~~~
+
+
+
+
+
+## Design Lowpass FIR filter
+
+
+Designing a lowpass FIR filter is very simple to do with DSP.jl, all you
+need to do is to define the window length, cut off frequency and the
+window. We will define a lowpass filter with cut off frequency at 5Hz for a signal
+sampled at 20 Hz.
+We will use the Hamming window, which is defined as:
+$w(n) = \alpha - \beta\cos\frac{2\pi n}{N-1}$, where $\alpha=0.54$ and $\beta=0.46$
+
+~~~~{.julia}
+fs = 20
+f = digitalfilter(Lowpass(5, fs = fs), FIRWindow(hamming(61)))
+w = range(0, stop=pi, length=1024)
+h = FIRfreqz(f, w)
+~~~~~~~~~~~~~
+
+
+~~~~
+1024-element Array{Complex{Float32},1}:
+ 1.0f0 + 0.0f0im
+ 0.99546844f0 + 0.095055714f0im
+ 0.98191506f0 + 0.1892486f0im
+ 0.95946306f0 + 0.28172377f0im
+ 0.9283168f0 + 0.37164196f0im
+ 0.8887594f0 + 0.45818728f0im
+ 0.84115064f0 + 0.54057467f0im
+ 0.7859234f0 + 0.618057f0im
+ 0.72357976f0 + 0.6899319f0im
+ 0.65468615f0 + 0.7555481f0im
+ ⋮
+ 0.00043952762f0 - 0.00041908873f0im
+ 0.0005152718f0 - 0.00040521423f0im
+ 0.0005873293f0 - 0.00037745363f0im
+ 0.0006531789f0 - 0.0003367371f0im
+ 0.0007105166f0 - 0.00028444792f0im
+ 0.0007573364f0 - 0.00022237403f0im
+ 0.0007920005f0 - 0.00015264557f0im
+ 0.0008132961f0 - 7.766036f-5im
+ 0.0008204784f0 - 3.1148685f-18im
+~~~~
+
+
+
+
+
+## Plot the frequency and impulse response
+
+
+The next code chunk is executed in term mode, see the [script](FIR_design.jl) for syntax.
+
+~~~~{.julia}
+julia> h_db = log10.(abs.(h));
+
+julia> ws = w/pi*(fs/2)
+0.0:0.009775171065493646:10.0
+
+~~~~~~~~~~~~~
+
+
+
+~~~~{.julia}
+plot(ws, h_db,
+ xlabel = "Frequency (Hz)", ylabel = "Magnitude (db)")
+~~~~~~~~~~~~~
+
+
+\
+
+
+
+
+And again with default options
+
+~~~~{.julia}
+h_phase = unwrap(-atan.(imag.(h),real.(h)))
+plot(ws, h_phase,
+ xlabel = "Frequency (Hz)", ylabel = "Phase (radians)")
+~~~~~~~~~~~~~
+
+
+\
+
diff --git a/v0.8.1/examples/FIR_design.txt b/v0.8.1/examples/FIR_design.txt
new file mode 100644
index 0000000..d9cae50
--- /dev/null
+++ b/v0.8.1/examples/FIR_design.txt
@@ -0,0 +1,152 @@
+---
+title: FIR filter design with Julia
+author: Matti Pastell
+date: 21th April 2016
+---
+
+
+# Introduction
+
+
+This an example of a julia script that can be published using
+[Weave](http://mpastell.github.io/Weave.jl/latest/usage/).
+The script can be executed normally using Julia
+or published to HTML or pdf with Weave.
+Text is written in markdown in lines starting with "`#'` " and code
+is executed and results are included in the published document.
+
+
+Notice that you don't need to define chunk options, but you can using
+`#+`. just before code e.g. `#+ term=True, caption='Fancy plots.'`.
+If you're viewing the published version have a look at the
+[source](FIR_design_plots.jl) to see the markup.
+
+
+
+# FIR Filter Design
+
+
+We'll implement lowpass, highpass and ' bandpass FIR filters. If
+you want to read more about DSP I highly recommend [The Scientist
+and Engineer's Guide to Digital Signal
+Processing](http://www.dspguide.com/) which is freely available
+online.
+
+
+## Calculating frequency response
+
+
+DSP.jl package doesn't (yet) have a method to calculate the
+the frequency response of a FIR filter so we define it:
+
+~~~~{.julia}
+using Plots, DSP
+gr()
+
+function FIRfreqz(b::Array, w = range(0, stop=π, length=1024))
+ n = length(w)
+ h = Array{ComplexF32}(undef, n)
+ sw = 0
+ for i = 1:n
+ for j = 1:length(b)
+ sw += b[j]*exp(-im*w[i])^-j
+ end
+ h[i] = sw
+ sw = 0
+ end
+ return h
+end
+~~~~~~~~~~~~~
+
+
+~~~~
+FIRfreqz (generic function with 2 methods)
+~~~~
+
+
+
+
+
+## Design Lowpass FIR filter
+
+
+Designing a lowpass FIR filter is very simple to do with DSP.jl, all you
+need to do is to define the window length, cut off frequency and the
+window. We will define a lowpass filter with cut off frequency at 5Hz for a signal
+sampled at 20 Hz.
+We will use the Hamming window, which is defined as:
+$w(n) = \alpha - \beta\cos\frac{2\pi n}{N-1}$, where $\alpha=0.54$ and $\beta=0.46$
+
+~~~~{.julia}
+fs = 20
+f = digitalfilter(Lowpass(5, fs = fs), FIRWindow(hamming(61)))
+w = range(0, stop=pi, length=1024)
+h = FIRfreqz(f, w)
+~~~~~~~~~~~~~
+
+
+~~~~
+1024-element Array{Complex{Float32},1}:
+ 1.0f0 + 0.0f0im
+ 0.99546844f0 + 0.095055714f0im
+ 0.98191506f0 + 0.1892486f0im
+ 0.95946306f0 + 0.28172377f0im
+ 0.9283168f0 + 0.37164196f0im
+ 0.8887594f0 + 0.45818728f0im
+ 0.84115064f0 + 0.54057467f0im
+ 0.7859234f0 + 0.618057f0im
+ 0.72357976f0 + 0.6899319f0im
+ 0.65468615f0 + 0.7555481f0im
+ ⋮
+ 0.00043952762f0 - 0.00041908873f0im
+ 0.0005152718f0 - 0.00040521423f0im
+ 0.0005873293f0 - 0.00037745363f0im
+ 0.0006531789f0 - 0.0003367371f0im
+ 0.0007105166f0 - 0.00028444792f0im
+ 0.0007573364f0 - 0.00022237403f0im
+ 0.0007920005f0 - 0.00015264557f0im
+ 0.0008132961f0 - 7.766036f-5im
+ 0.0008204784f0 - 3.1148685f-18im
+~~~~
+
+
+
+
+
+## Plot the frequency and impulse response
+
+
+The next code chunk is executed in term mode, see the [script](FIR_design.jl) for syntax.
+
+~~~~{.julia}
+julia> h_db = log10.(abs.(h));
+
+julia> ws = w/pi*(fs/2)
+0.0:0.009775171065493646:10.0
+
+~~~~~~~~~~~~~
+
+
+
+~~~~{.julia}
+plot(ws, h_db,
+ xlabel = "Frequency (Hz)", ylabel = "Magnitude (db)")
+~~~~~~~~~~~~~
+
+
+\
+
+
+
+
+And again with default options
+
+~~~~{.julia}
+h_phase = unwrap(-atan.(imag.(h),real.(h)))
+plot(ws, h_phase,
+ xlabel = "Frequency (Hz)", ylabel = "Phase (radians)")
+~~~~~~~~~~~~~
+
+
+\
+
diff --git a/v0.8.1/examples/FIR_design_plots.html b/v0.8.1/examples/FIR_design_plots.html
new file mode 100644
index 0000000..fa16a24
--- /dev/null
+++ b/v0.8.1/examples/FIR_design_plots.html
@@ -0,0 +1,789 @@
+
+
+
+
+
+ FIR filter design with Julia
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ FIR filter design with Julia
+ Matti Pastell
+ 21th April 2016
+
+
+
+Introduction
+
+This an example of a julia script that can be published using Weave. The script can be executed normally using Julia or published to HTML or pdf with Weave. Text is written in markdown in lines starting with "#' " and code is executed and results are included in the published document.
+
+Notice that you don't need to define chunk options, but you can using #+. just before code e.g. #+ term=True, caption='Fancy plots.'. If you're viewing the published version have a look at the source to see the markup.
+
+FIR Filter Design
+
+We'll implement lowpass, highpass and ' bandpass FIR filters. If you want to read more about DSP I highly recommend The Scientist and Engineer's Guide to Digital Signal Processing which is freely available online.
+
+Calculating frequency response
+
+DSP.jl package doesn't (yet) have a method to calculate the the frequency response of a FIR filter so we define it:
+
+
+
+using Plots, DSP
+gr()
+
+function FIRfreqz(b::Array, w = range(0, stop=π, length=1024))
+ n = length(w)
+ h = Array{ComplexF32}(undef, n)
+ sw = 0
+ for i = 1:n
+ for j = 1:length(b)
+ sw += b[j]*exp(-im*w[i])^-j
+ end
+ h[i] = sw
+ sw = 0
+ end
+ return h
+end
+
+
+
+
+FIRfreqz (generic function with 2 methods)
+
+
+
+Design Lowpass FIR filter
+
+Designing a lowpass FIR filter is very simple to do with DSP.jl, all you need to do is to define the window length, cut off frequency and the window. We will define a lowpass filter with cut off frequency at 5Hz for a signal sampled at 20 Hz. We will use the Hamming window, which is defined as: $w(n) = \alpha - \beta\cos\frac{2\pi n}{N-1}$, where $\alpha=0.54$ and $\beta=0.46$
+
+
+
+fs = 20
+f = digitalfilter(Lowpass(5, fs = fs), FIRWindow(hamming(61)))
+w = range(0, stop=pi, length=1024)
+h = FIRfreqz(f, w)
+
+
+
+
+1024-element Array{Complex{Float32},1}:
+ 1.0f0 + 0.0f0im
+ 0.99546844f0 + 0.095055714f0im
+ 0.98191506f0 + 0.1892486f0im
+ 0.95946306f0 + 0.28172377f0im
+ 0.9283168f0 + 0.37164196f0im
+ 0.8887594f0 + 0.45818728f0im
+ 0.84115064f0 + 0.54057467f0im
+ 0.7859234f0 + 0.618057f0im
+ 0.72357976f0 + 0.6899319f0im
+ 0.65468615f0 + 0.7555481f0im
+ ⋮
+ 0.00043952762f0 - 0.00041908873f0im
+ 0.0005152718f0 - 0.00040521423f0im
+ 0.0005873293f0 - 0.00037745363f0im
+ 0.0006531789f0 - 0.0003367371f0im
+ 0.0007105166f0 - 0.00028444792f0im
+ 0.0007573364f0 - 0.00022237403f0im
+ 0.0007920005f0 - 0.00015264557f0im
+ 0.0008132961f0 - 7.766036f-5im
+ 0.0008204784f0 - 3.1148685f-18im
+
+
+
+Plot the frequency and impulse response
+
+The next code chunk is executed in term mode, see the script for syntax.
+
+
+julia> h_db = log10.(abs.(h));
+
+julia> ws = w/pi*(fs/2)
+0.0:0.009775171065493646:10.0
+
+
+
+
+plot(ws, h_db,
+ xlabel = "Frequency (Hz)", ylabel = "Magnitude (db)")
+
+
+
+ +
+
+
+And again with default options
+
+
+
+h_phase = unwrap(-atan.(imag.(h),real.(h)))
+plot(ws, h_phase,
+ xlabel = "Frequency (Hz)", ylabel = "Phase (radians)")
+
+
+
+ +
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
diff --git a/v0.8.1/examples/FIR_design_plots.jl b/v0.8.1/examples/FIR_design_plots.jl
new file mode 100644
index 0000000..75844b1
--- /dev/null
+++ b/v0.8.1/examples/FIR_design_plots.jl
@@ -0,0 +1,85 @@
+#' ---
+#' title: FIR filter design with Julia
+#' author: Matti Pastell
+#' date: 21th April 2016
+#' ---
+
+#' # Introduction
+
+#' This an example of a julia script that can be published using
+#' [Weave](http://mpastell.github.io/Weave.jl/latest/usage/).
+#' The script can be executed normally using Julia
+#' or published to HTML or pdf with Weave.
+#' Text is written in markdown in lines starting with "`#'` " and code
+#' is executed and results are included in the published document.
+
+#' Notice that you don't need to define chunk options, but you can using
+#' `#+`. just before code e.g. `#+ term=True, caption='Fancy plots.'`.
+#' If you're viewing the published version have a look at the
+#' [source](FIR_design_plots.jl) to see the markup.
+
+
+#' # FIR Filter Design
+
+#' We'll implement lowpass, highpass and ' bandpass FIR filters. If
+#' you want to read more about DSP I highly recommend [The Scientist
+#' and Engineer's Guide to Digital Signal
+#' Processing](http://www.dspguide.com/) which is freely available
+#' online.
+
+#' ## Calculating frequency response
+
+#' DSP.jl package doesn't (yet) have a method to calculate the
+#' the frequency response of a FIR filter so we define it:
+
+using Plots, DSP
+gr()
+
+function FIRfreqz(b::Array, w = range(0, stop=π, length=1024))
+ n = length(w)
+ h = Array{ComplexF32}(undef, n)
+ sw = 0
+ for i = 1:n
+ for j = 1:length(b)
+ sw += b[j]*exp(-im*w[i])^-j
+ end
+ h[i] = sw
+ sw = 0
+ end
+ return h
+end
+
+
+#' ## Design Lowpass FIR filter
+
+#' Designing a lowpass FIR filter is very simple to do with DSP.jl, all you
+#' need to do is to define the window length, cut off frequency and the
+#' window. We will define a lowpass filter with cut off frequency at 5Hz for a signal
+#' sampled at 20 Hz.
+#' We will use the Hamming window, which is defined as:
+#' $w(n) = \alpha - \beta\cos\frac{2\pi n}{N-1}$, where $\alpha=0.54$ and $\beta=0.46$
+
+
+fs = 20
+f = digitalfilter(Lowpass(5, fs = fs), FIRWindow(hamming(61)))
+w = range(0, stop=pi, length=1024)
+h = FIRfreqz(f, w)
+
+#' ## Plot the frequency and impulse response
+
+#' The next code chunk is executed in term mode, see the [script](FIR_design.jl) for syntax.
+#+ term=true
+
+h_db = log10.(abs.(h));
+ws = w/pi*(fs/2)
+
+#+
+
+plot(ws, h_db,
+ xlabel = "Frequency (Hz)", ylabel = "Magnitude (db)")
+
+#' And again with default options
+
+h_phase = unwrap(-atan.(imag.(h),real.(h)))
+plot(ws, h_phase,
+ xlabel = "Frequency (Hz)", ylabel = "Phase (radians)")
diff --git a/v0.8.1/examples/figures/FIR_design_4_1.png b/v0.8.1/examples/figures/FIR_design_4_1.png
new file mode 100644
index 0000000..24616a7
Binary files /dev/null and b/v0.8.1/examples/figures/FIR_design_4_1.png differ
diff --git a/v0.8.1/examples/figures/FIR_design_5_1.png b/v0.8.1/examples/figures/FIR_design_5_1.png
new file mode 100644
index 0000000..76e263e
Binary files /dev/null and b/v0.8.1/examples/figures/FIR_design_5_1.png differ
diff --git a/v0.8.1/function_index/index.html b/v0.8.1/function_index/index.html
new file mode 100644
index 0000000..5785d08
--- /dev/null
+++ b/v0.8.1/function_index/index.html
@@ -0,0 +1,2 @@
+
+Function index · Weave.jl
Function index
Getting started · Weave.jl
Getting started
The best way to get started using Weave.jl is to look at the example input and output documents. Examples for different formats are included in the packages examples directory.
First have a look at source document using markdown code chunks and Plots.jl for figures: FIR_design.jmd and then see the output in different formats:
- HTML: FIR_design.html
- pdf: FIR_design.pdf
- Pandoc markdown: FIR_design.txt
Producing pdf output requires that you have XeLateX installed.
Add dependencies for the example if needed:
using Pkg; Pkg.add.(["Plots", "DSP"])
Weave the files to your working directory using:
using Weave
+#HTML
+weave(joinpath(dirname(pathof(Weave)), "../examples", "FIR_design.jmd"),
+ out_path=:pwd,
+ doctype = "md2html")
+#pdf
+weave(joinpath(dirname(pathof(Weave)), "../examples", "FIR_design.jmd"),
+ out_path=:pwd,
+ doctype = "md2pdf")
+ #Markdown
+weave(joinpath(dirname(pathof(Weave)), "../examples", "FIR_design.jmd"),
+ doctype="pandoc"
+ out_path=:pwd)
Weave.jl - Scientific Reports Using Julia · Weave.jl
Weave.jl - Scientific Reports Using Julia
This is the documentation of Weave.jl. Weave is a scientific report generator/literate programming tool for Julia. It resembles Pweave, Knitr, rmarkdown and Sweave.
Current features
- Markdown, script of Noweb syntax for input documents.
- Publish markdown directly to html and pdf using Julia or Pandoc markdown.
- Execute code as terminal or "script" chunks.
- Capture Plots.jl or Gadfly.jl figures
- Supports LaTex, Pandoc, Github markdown, MultiMarkdown, Asciidoc and reStructuredText output
- Simple caching of results
- Convert to and from IJulia notebooks
Contents
Working with Jupyter notebooks · Weave.jl
Working with Jupyter notebooks
Weaving from Jupyter notebooks
Weave supports using Jupyter notebooks as input format, this means you can weave notebooks to any supported formats. You can't use chunk options with notebooks.
weave("notebook.ipynb")
Output to Jupyter notebooks
As of Weave 0.5.1. there is new notebook method to convert Weave documents to Jupyter notebooks using nbconvert. The code is not executed by Weave and the output doesn't always work properly, see #116.
Weave.notebook — Function.notebook(source::String, outpath=:pwd, timeout=-1, nbconvertoptions="")
Convert Weave document source to Jupyter notebook and execute the code using nbconvert. Requires IJulia. Ignores all chunk options
out_path: Path where the output is generated. Can be: :doc: Path of the source document, :pwd: Julia working directory, "somepath": Path as a String e.g "/home/mpastell/weaveout"timeout: nbconvert cell timeout in seconds. Defaults to -1 (no timeout)nbconvert_options: string of additional options to pass to nbconvert, such as --allow-errors
sourceYou might wan't to use the convert_doc method below instead and run the code in Jupyter.
Converting between formats
You can convert between all supported input formats using the convert_doc function.
To convert from script to notebook:
convert_doc("examples/FIR_design.jl", "FIR_design.ipynb")
and from notebooks to markdown use:
convert_doc("FIR_design.ipynb", "FIR_design.jmd")
Weave.convert_doc — Method.convert_doc(infile::AbstractString, outfile::AbstractString; format = nothing)
Convert Weave documents between different formats
infile = Name of the input documentoutfile = Name of the output documentformat = Output format (optional). Detected from outfile extension, but can be set to "script", "markdown", "notebook" or "noweb".
sourcePublishing to html and pdf · Weave.jl
Publishing to html and pdf
You can also publish any supported input format using markdown for doc chunks to html and pdf documents. Producing pdf output requires that you have pdflatex installed and in your path.
You can use a YAML header in the beginning of the input document delimited with "–-" to set the document title, author and date e.g.
---
+title : Weave example
+author : Matti Pastell
+date: 15th December 2016
+---
Here is a a sample document and output:
FIRdesignplots.jl, FIRdesignplots.html , FIRdesignplots.pdf.
weave("FIR_design_plots.jl")
+weave("FIR_design_plots.jl", docformat = "md2pdf")
Note: docformats md2pdf and md2html use Julia markdown and pandoc2pdf and pandoc2html use Pandoc.
Templates
You can use a custom template with md2pdf and md2html formats with template argument (e.g) weave("FIR_design_plots.jl", template = "custom.tpl"). You can use the existing templates as starting point.
For HTML: julia_html.tpl and LaTex: julia_tex.tpl
Templates are rendered using Mustache.jl.
Supported Markdown syntax
The markdown variant used by Weave is Julia markdown. In addition Weave supports few additional Markdown features:
Comments
You can add comments using html syntax: <!-- -->
Multiline equations
You can add multiline equations using:
$$
+x^2 = x*x
+$$
Search · Weave.jl
Search
Number of results: loading...
>= and end with line marked with @. The code between the start and end markers is executed and the output is captured to the output document. See chunk options."
+},
+
+{
+ "location": "usage/#Script-format-1",
+ "page": "Using Weave",
+ "title": "Script format",
+ "category": "section",
+ "text": "Weave also support script input format with a markup in comments. These scripts can be executed normally using Julia or published with Weave. Documentation is in lines starting with #\', #%% or # %%, and code is executed and results are included in the weaved document.All lines that are not documentation are treated as code. You can set chunk options using lines starting with #+ just before code e.g. #+ term=true.The format is identical to Pweave and the concept is similar to publishing documents with MATLAB or using Knitr\'s spin. Weave will remove the first empty space from each line of documentation.See sample document:"
+},
+
+{
+ "location": "usage/#Inline-code-1",
+ "page": "Using Weave",
+ "title": "Inline code",
+ "category": "section",
+ "text": "You can also add inline code to your documents using`j juliacode`syntax. The code will be replaced with the output of running the code. If the code produces figures the filename or base64 encoded string will be added to output e.g. to include a Plots figure in markdown you can use:`)"
+},
+
+{
+ "location": "usage/#Setting-document-options-in-header-1",
+ "page": "Using Weave",
+ "title": "Setting document options in header",
+ "category": "section",
+ "text": "You can use a YAML header in the beginning of the input document delimited with \"–-\" to set the document title, author and date e.g. and default document options. Each of Weave command line arguments and chunk options can be set in header using options field. Below is an example that sets document out_path and doctype using the header.---\ntitle : Weave example\nauthor : Matti Pastell\ndate: 15th December 2016\noptions:\n out_path : reports/example.md\n doctype : github\n---You can also set format specific options. Here is how to set different outpath for md2html and md2pdf and set `figext` for both:---\noptions:\n md2html:\n out_path : html\n md2pdf:\n out_path : pdf\n fig_ext : .png\n---"
+},
+
+{
+ "location": "usage/#Passing-arguments-to-documents-1",
+ "page": "Using Weave",
+ "title": "Passing arguments to documents",
+ "category": "section",
+ "text": "You can pass arguments as dictionary to the weaved document using the args argument to weave. The dictionary will be available as WEAVE_ARGS variable in the document.This makes it possible to create the same report easily for e.g. different date ranges of input data from a database or from files with similar format giving the filename as input.In order to pass a filename to a document you need call weave using:weave(\"mydoc.jmd\", args = Dict(\"filename\" => \"somedata.h5\"))and you can access the filename from document as follows: ```julia\n print(WEAVE_ARGS[\"filename\"])\n ```You can use the out_path argument to control the name of the output document."
+},
+
+{
+ "location": "usage/#Weave.include_weave",
+ "page": "Using Weave",
+ "title": "Weave.include_weave",
+ "category": "function",
+ "text": "include_weave(doc, informat=:auto)\n\nInclude code from Weave document calling include_string on all code from doc. Code is run in the path of the include document.\n\n\n\n\n\n"
+},
+
+{
+ "location": "usage/#Include-Weave-document-in-Julia-1",
+ "page": "Using Weave",
+ "title": "Include Weave document in Julia",
+ "category": "section",
+ "text": "You can call include_weave on a Weave document to run the contents of all code chunks in Julia.include_weave(doc, informat=:auto)"
+},
+
+{
+ "location": "publish/#",
+ "page": "Publishing to html and pdf",
+ "title": "Publishing to html and pdf",
+ "category": "page",
+ "text": ""
+},
+
+{
+ "location": "publish/#Publishing-to-html-and-pdf-1",
+ "page": "Publishing to html and pdf",
+ "title": "Publishing to html and pdf",
+ "category": "section",
+ "text": "You can also publish any supported input format using markdown for doc chunks to html and pdf documents. Producing pdf output requires that you have pdflatex installed and in your path.You can use a YAML header in the beginning of the input document delimited with \"–-\" to set the document title, author and date e.g.---\ntitle : Weave example\nauthor : Matti Pastell\ndate: 15th December 2016\n---Here is a a sample document and output:FIRdesignplots.jl, FIRdesignplots.html , FIRdesignplots.pdf.weave(\"FIR_design_plots.jl\")\nweave(\"FIR_design_plots.jl\", docformat = \"md2pdf\")Note: docformats md2pdf and md2html use Julia markdown and pandoc2pdf and pandoc2html use Pandoc."
+},
+
+{
+ "location": "publish/#Templates-1",
+ "page": "Publishing to html and pdf",
+ "title": "Templates",
+ "category": "section",
+ "text": "You can use a custom template with md2pdf and md2html formats with template argument (e.g) weave(\"FIR_design_plots.jl\", template = \"custom.tpl\"). You can use the existing templates as starting point.For HTML: julia_html.tpl and LaTex: julia_tex.tplTemplates are rendered using Mustache.jl."
+},
+
+{
+ "location": "publish/#Supported-Markdown-syntax-1",
+ "page": "Publishing to html and pdf",
+ "title": "Supported Markdown syntax",
+ "category": "section",
+ "text": "The markdown variant used by Weave is Julia markdown. In addition Weave supports few additional Markdown features:CommentsYou can add comments using html syntax: Multiline equationsYou can add multiline equations using:$$\nx^2 = x*x\n$$"
+},
+
+{
+ "location": "chunk_options/#",
+ "page": "Chunk options",
+ "title": "Chunk options",
+ "category": "page",
+ "text": ""
+},
+
+{
+ "location": "chunk_options/#Chunk-options-1",
+ "page": "Chunk options",
+ "title": "Chunk options",
+ "category": "section",
+ "text": "I\'ve mostly followed Knitr\'s naming for chunk options, but not all options are implemented.Options are separated using \";\" and need to be valid Julia expressions. Example: markdown code chunk that saves and displays a 12 cm wide image and hides the source code:julia; out_width=\"12cm\"; echo=falseWeave currently supports the following chunk options with the following defaults:"
+},
+
+{
+ "location": "chunk_options/#Options-for-code-1",
+ "page": "Chunk options",
+ "title": "Options for code",
+ "category": "section",
+ "text": "echo = true. Echo the code in the output document. If false the source code will be hidden.\nresults = \"markup\". The output format of the printed results. \"markup\" for literal block, \"hidden\" for hidden results or anything else for raw output (I tend to use ‘tex’ for Latex and ‘rst’ for rest. Raw output is useful if you wan’t to e.g. create tables from code chunks.\neval = true. Evaluate the code chunk. If false the chunk won’t be executed.\nterm=false. If true the output emulates a REPL session. Otherwise only stdout and figures will be included in output.\nlabel. Chunk label, will be used for figure labels in Latex as fig:label\nwrap = true. Wrap long lines from output.\nline_width = 75. Line width for wrapped lines.\ncache = false. Cache results, depends on cache parameter on weave function.\nhold = false. Hold all results until the end of the chunk.\ntangle = true. Set tangle to false to exclude chunk from tangled code."
+},
+
+{
+ "location": "chunk_options/#Options-for-figures-1",
+ "page": "Chunk options",
+ "title": "Options for figures",
+ "category": "section",
+ "text": "fig_width. Figure width passed to plotting library e.g. 800\nfig_height Figure height passed to plotting library\nout_width. Width of saved figure in output markup e.g. \"50%\", \"12cm\", \\\\0.5linewidth\nout_height. Height of saved figure in output markup\ndpi=96. Resolution of saved figures.\nfig_cap. Figure caption.\nlabel. Chunk label, will be used for figure labels in Latex as fig:label\nfig_ext. File extension (format) of saved figures.\nfig_pos=\"htpb\". Figure position in Latex. \nfig_env=\"figure\". Figure environment in Latex."
+},
+
+{
+ "location": "chunk_options/#Set-default-chunk-options-1",
+ "page": "Chunk options",
+ "title": "Set default chunk options",
+ "category": "section",
+ "text": "You can set the default chunk options (and weave arguments) for a document using the YAML header options field. e.g to set the default out_width of all figures you can use:---\noptions:\n out_width : 50%\n---You can also set or change the default chunk options for a document either before weave using the set_chunk_defaults function.set_chunk_defaults(opts)\nget_chunk_defaults()\nrestore_chunk_defaults()"
+},
+
+{
+ "location": "notebooks/#",
+ "page": "Working with Jupyter notebooks",
+ "title": "Working with Jupyter notebooks",
+ "category": "page",
+ "text": ""
+},
+
+{
+ "location": "notebooks/#Working-with-Jupyter-notebooks-1",
+ "page": "Working with Jupyter notebooks",
+ "title": "Working with Jupyter notebooks",
+ "category": "section",
+ "text": ""
+},
+
+{
+ "location": "notebooks/#Weaving-from-Jupyter-notebooks-1",
+ "page": "Working with Jupyter notebooks",
+ "title": "Weaving from Jupyter notebooks",
+ "category": "section",
+ "text": "Weave supports using Jupyter notebooks as input format, this means you can weave notebooks to any supported formats. You can\'t use chunk options with notebooks.weave(\"notebook.ipynb\")"
+},
+
+{
+ "location": "notebooks/#Weave.notebook",
+ "page": "Working with Jupyter notebooks",
+ "title": "Weave.notebook",
+ "category": "function",
+ "text": "notebook(source::String, outpath=:pwd, timeout=-1, nbconvertoptions=\"\")\n\nConvert Weave document source to Jupyter notebook and execute the code using nbconvert. Requires IJulia. Ignores all chunk options\n\nout_path: Path where the output is generated. Can be: :doc: Path of the source document, :pwd: Julia working directory, \"somepath\": Path as a String e.g \"/home/mpastell/weaveout\"\ntimeout: nbconvert cell timeout in seconds. Defaults to -1 (no timeout)\nnbconvert_options: string of additional options to pass to nbconvert, such as --allow-errors\n\n\n\n\n\n"
+},
+
+{
+ "location": "notebooks/#Output-to-Jupyter-notebooks-1",
+ "page": "Working with Jupyter notebooks",
+ "title": "Output to Jupyter notebooks",
+ "category": "section",
+ "text": "As of Weave 0.5.1. there is new notebook method to convert Weave documents to Jupyter notebooks using nbconvert. The code is not executed by Weave and the output doesn\'t always work properly, see #116.notebook(source::String, out_path=:pwd)You might wan\'t to use the convert_doc method below instead and run the code in Jupyter."
+},
+
+{
+ "location": "notebooks/#Weave.convert_doc-Tuple{String,String}",
+ "page": "Working with Jupyter notebooks",
+ "title": "Weave.convert_doc",
+ "category": "method",
+ "text": "convert_doc(infile::AbstractString, outfile::AbstractString; format = nothing)\n\nConvert Weave documents between different formats\n\ninfile = Name of the input document\noutfile = Name of the output document\nformat = Output format (optional). Detected from outfile extension, but can be set to \"script\", \"markdown\", \"notebook\" or \"noweb\".\n\n\n\n\n\n"
+},
+
+{
+ "location": "notebooks/#Converting-between-formats-1",
+ "page": "Working with Jupyter notebooks",
+ "title": "Converting between formats",
+ "category": "section",
+ "text": "You can convert between all supported input formats using the convert_doc function.To convert from script to notebook:convert_doc(\"examples/FIR_design.jl\", \"FIR_design.ipynb\")and from notebooks to markdown use:convert_doc(\"FIR_design.ipynb\", \"FIR_design.jmd\")convert_doc(infile::String, outfile::String)"
+},
+
+{
+ "location": "function_index/#",
+ "page": "Function index",
+ "title": "Function index",
+ "category": "page",
+ "text": ""
+},
+
+{
+ "location": "function_index/#Function-index-1",
+ "page": "Function index",
+ "title": "Function index",
+ "category": "section",
+ "text": ""
+},
+
+]}
diff --git a/v0.8.1/siteinfo.js b/v0.8.1/siteinfo.js
new file mode 100644
index 0000000..a6be1ae
--- /dev/null
+++ b/v0.8.1/siteinfo.js
@@ -0,0 +1 @@
+var DOCUMENTER_CURRENT_VERSION = "v0.8.1";
diff --git a/v0.8.1/usage/index.html b/v0.8.1/usage/index.html
new file mode 100644
index 0000000..2c2c6e6
--- /dev/null
+++ b/v0.8.1/usage/index.html
@@ -0,0 +1,48 @@
+
+Using Weave · Weave.jl
Using Weave
You can write your documentation and code in input document using Markdown, Noweb or script syntax and use weave function to execute to document to capture results and figures.
Weave
Weave document with markup and julia code using Plots.jl for plots, out_path = :pwd makes the results appear in the current working directory.
#First add depencies for the example
+using Pkg; Pkg.add.(["Plots", "DSP"])
+using Weave
+weave(joinpath(dirname(pathof(Weave)), "../examples", "FIR_design.jmd"), out_path=:pwd)
Weave.weave — Method.weave(source ; doctype = :auto,
+ informat=:auto, out_path=:doc, args = Dict(),
+ mod::Union{Module, Symbol} = Main,
+ fig_path = "figures", fig_ext = nothing,
+ cache_path = "cache", cache=:off,
+ template = nothing, highlight_theme = nothing, css = nothing,
+ pandoc_options = "",
+ latex_cmd = "xelatex")
Weave an input document to output file.
doctype: :auto = set based on file extension or specify one of the supported formats. See list_out_formats()informat: :auto = set based on file extension or set to "noweb", "markdown" or scriptout_path: Path where the output is generated. Can be: :doc: Path of the source document, :pwd: Julia working directory, "somepath": output directory as a String e.g "/home/mpastell/weaveout" or filename as string e.g. ~/outpath/outfile.tex.args: dictionary of arguments to pass to document. Available as WEAVE_ARGSmod: Module where Weave evals code. Defaults to :sandbox to create new sandbox module, you can also pass a module e.g. Main.fig_path: where figures will be generated, relative to out_pathfig_ext: Extension for saved figures e.g. ".pdf", ".png". Default setting depends on doctype.cache_path: where of cached output will be saved.cache: controls caching of code: :off = no caching, :all = cache everything, :user = cache based on chunk options, :refresh, run all code chunks and save new cache.throw_errors if false errors are included in output document and the whole document is executed. if true errors are thrown when they occur.template : Template (file path) for md2html or md2tex formats.highlight_theme : Theme (Highlights.AbstractTheme) for used syntax highlightingcss : CSS (file path) used for md2html formatpandoc_options = String array of options to pass to pandoc for pandoc2html and pandoc2pdf formats e.g. ["–toc", "-N"]latex_cmd the command used to make pdf from .tex
Note: Run Weave from terminal and not using IJulia, Juno or ESS, they tend to mess with capturing output.
sourceTangle
Tangling extracts the code from document:
Weave.tangle — Method.tangle(source ; out_path=:doc, informat="noweb")
Tangle source code from input document to .jl file.
informat: "noweb" of "markdown"out_path: Path where the output is generated. Can be: :doc: Path of the source document, :pwd: Julia working directory, "somepath", directory name as a string e.g "/home/mpastell/weaveout"
or filename as string e.g. ~/outpath/outfile.jl.
sourceSupported output formats
Weave sets the output format based on the file extension, but you can also set it using doctype option. The rules for detecting the format are:
ext == ".jl" && return "md2html"
+contains(ext, ".md") && return "md2html"
+contains(ext, ".rst") && return "rst"
+contains(ext, ".tex") && return "texminted"
+contains(ext, ".txt") && return "asciidoc"
+return "pandoc"
You can get a list of supported output formats:
list_out_formats()
github: Github markdown
+md2tex: Julia markdown to latex
+pandoc2html: Markdown to HTML (requires Pandoc 2)
+pandoc: Pandoc markdown
+pandoc2pdf: Pandoc markdown
+tex: Latex with custom code environments
+texminted: Latex using minted for highlighting
+md2html: Julia markdown to html
+rst: reStructuredText and Sphinx
+multimarkdown: MultiMarkdown
+md2pdf: Julia markdown to latex
+asciidoc: AsciiDoc
+hugo: Hugo markdown (using shortcodes)
Weave.list_out_formats — Method.list_out_formats()
List supported output formats
sourceDocument syntax
Weave uses markdown, Noweb or script syntax for defining the code chunks and documentation chunks. You can also weave Jupyter notebooks. The format is detected based on the file extension, but you can also set it manually using the informat parameter.
The rules for autodetection are:
ext == ".jl" && return "script"
+ext == ".jmd" && return "markdown"
+ext == ".ipynb" && return "notebook"
+return "noweb"
Documentation chunks
In Markdown and Noweb input formats documentation chunks are the parts that aren't inside code delimiters. Documentation chunks can be written with several different markup languages.
Code chunks
Markdown format
Markdown code chunks are defined using fenced code blocks with options following on the same line. e.g. to hide code from output you can use:
julia; echo=false
Noweb format
Code chunks start with a line marked with <<>>= or <<options>>= and end with line marked with @. The code between the start and end markers is executed and the output is captured to the output document. See chunk options.
Script format
Weave also support script input format with a markup in comments. These scripts can be executed normally using Julia or published with Weave. Documentation is in lines starting with #', #%% or # %%, and code is executed and results are included in the weaved document.
All lines that are not documentation are treated as code. You can set chunk options using lines starting with #+ just before code e.g. #+ term=true.
The format is identical to Pweave and the concept is similar to publishing documents with MATLAB or using Knitr's spin. Weave will remove the first empty space from each line of documentation.
Inline code
You can also add inline code to your documents using
`j juliacode`
syntax. The code will be replaced with the output of running the code. If the code produces figures the filename or base64 encoded string will be added to output e.g. to include a Plots figure in markdown you can use:
`)
Setting document options in header
You can use a YAML header in the beginning of the input document delimited with "–-" to set the document title, author and date e.g. and default document options. Each of Weave command line arguments and chunk options can be set in header using options field. Below is an example that sets document out_path and doctype using the header.
---
+title : Weave example
+author : Matti Pastell
+date: 15th December 2016
+options:
+ out_path : reports/example.md
+ doctype : github
+---
You can also set format specific options. Here is how to set different outpath for md2html and md2pdf and set `figext` for both:
---
+options:
+ md2html:
+ out_path : html
+ md2pdf:
+ out_path : pdf
+ fig_ext : .png
+---
Passing arguments to documents
You can pass arguments as dictionary to the weaved document using the args argument to weave. The dictionary will be available as WEAVE_ARGS variable in the document.
This makes it possible to create the same report easily for e.g. different date ranges of input data from a database or from files with similar format giving the filename as input.
In order to pass a filename to a document you need call weave using:
weave("mydoc.jmd", args = Dict("filename" => "somedata.h5"))
and you can access the filename from document as follows:
```julia
+ print(WEAVE_ARGS["filename"])
+ ```
You can use the out_path argument to control the name of the output document.
Include Weave document in Julia
You can call include_weave on a Weave document to run the contents of all code chunks in Julia.
Weave.include_weave — Function.include_weave(doc, informat=:auto)
Include code from Weave document calling include_string on all code from doc. Code is run in the path of the include document.
source