mirror of https://github.com/mpastell/Weave.jl
Updated docs, added convert_doc
parent
15157a213a
commit
6a68be5161
|
|
@ -12,10 +12,11 @@ You can write your documentation and code in input document using Nowed or Markd
|
|||
|
||||
* Noweb, markdown or script syntax for input documents.
|
||||
* Execute code as terminal or "script" chunks.
|
||||
* Capture Gadfly, PyPlot and Winston figures.
|
||||
* Capture Plots, Gadfly, PyPlot and Winston figures.
|
||||
* Supports LaTex, Pandoc, Github markdown, MultiMarkdown, Asciidoc and reStructuredText output
|
||||
* Publish markdown directly to html and pdf using Pandoc.
|
||||
* Simple caching of results
|
||||
* Convert to and from IJulia notebooks
|
||||
|
||||
## Usage
|
||||
|
||||
|
|
|
|||
|
|
@ -31,4 +31,5 @@ pages:
|
|||
- Using Weave: usage.md
|
||||
- Publishing scripts: publish.md
|
||||
- Chunk options: chunk_options.md
|
||||
- Working with Jupyter notebooks: notebooks.md
|
||||
- Function index: function_index.md
|
||||
|
|
|
|||
|
|
@ -10,14 +10,18 @@ and Sweave.
|
|||
|
||||
* Noweb, markdown or script syntax for input documents.
|
||||
* Execute code as terminal or "script" chunks.
|
||||
* Capture Gadfly, PyPlot and Winston figures.
|
||||
* Capture Plots, Gadfly, PyPlot and Winston figures.
|
||||
* Supports LaTex, Pandoc, Github markdown, MultiMarkdown, Asciidoc and reStructuredText output
|
||||
* Publish markdown directly to html and pdf using Pandoc.
|
||||
* Simple caching of results
|
||||
* Convert to and from IJulia notebooks
|
||||
|
||||
|
||||
|
||||
## Contents
|
||||
|
||||
```@contents
|
||||
Pages = ["getting_started.md", "usage.md",
|
||||
"publish.md", "chunk_options.md", "notebooks.md",
|
||||
"function_index.md"]
|
||||
```
|
||||
|
|
|
|||
|
|
@ -0,0 +1,34 @@
|
|||
|
||||
# Working with Jupyter notebooks
|
||||
|
||||
## Weaving
|
||||
|
||||
Weave supports using Jupyter notebooks as input format, this means you can weave notebooks to any supported formats. You can't use chunk options with notebooks.
|
||||
|
||||
```
|
||||
weave("notebook.ipynb")
|
||||
```
|
||||
|
||||
In order to output notebooks from other formats you need to convert the
|
||||
document to a notebook and run the code using IJulia.
|
||||
|
||||
## Converting between formats
|
||||
|
||||
You can convert between all supported input formats using the `convert_doc`
|
||||
function.
|
||||
|
||||
To convert from script to notebook:
|
||||
|
||||
```
|
||||
convert_doc("examples/FIR_design.jl", "FIR_design.ipynb")
|
||||
```
|
||||
|
||||
and from notebooks to markdown use:
|
||||
|
||||
```
|
||||
convert_doc("FIR_design.ipynb", "FIR_design.jmd")
|
||||
```
|
||||
|
||||
```@docs
|
||||
convert_doc(infile::String, outfile::String)
|
||||
```
|
||||
|
|
@ -5,7 +5,7 @@ documents from Julia scripts with a specific format. Producing HTML and pdf outp
|
|||
requires that you have Pandoc and XeLatex (for pdf) installed and in your path.
|
||||
|
||||
These scripts can be executed normally using Julia or published with Weave.
|
||||
Documentation is written in markdown in lines starting with ``#'``, ``#%%`` or ``# %%`` ,
|
||||
Documentation is written in markdown in lines starting with `#'`, `#%%` or `# %%`,
|
||||
and code is executed and results are included in the published document.
|
||||
|
||||
The format is identical to [Pweave](http://mpastell.com/pweave/pypublish.html)
|
||||
|
|
@ -14,10 +14,9 @@ using Knitr's [spin](http://yihui.name/knitr/demo/stitch/).
|
|||
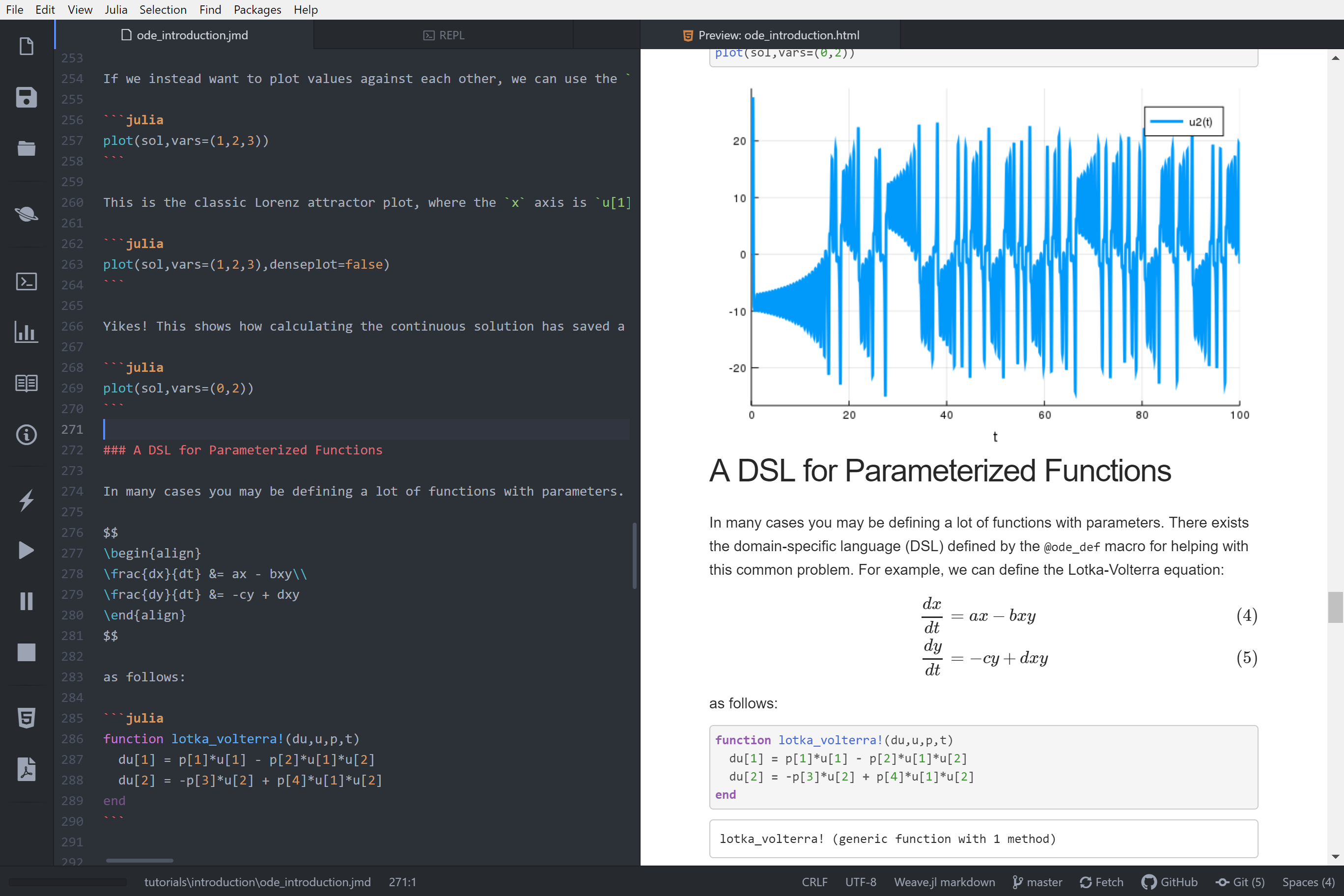
Weave will remove the first empty space from each line of documentation.
|
||||
|
||||
All lines that are not documentation are treated as code. You can set chunk options
|
||||
using lines starting with ``#+``, ``#%%`` or ``# %%`` just before code
|
||||
e.g. ``#+ term=True, caption='Fancy plots.'``. See the example below for the markup.
|
||||
using lines starting with `#+` just before code
|
||||
e.g. `#+ term=true`. See the example below for the markup.
|
||||
|
||||
The scripts can be published using the `pypublish` scipts:
|
||||
|
||||
[FIR_design.jl](examples/FIR_design.jl), [FIR_design.html](examples/FIR_design.html) , [FIR_design.pdf](examples/FIR_design.pdf).
|
||||
|
||||
|
|
|
|||
|
|
@ -78,19 +78,17 @@ list_out_formats()
|
|||
## Document syntax
|
||||
|
||||
Weave uses noweb, markdown or script syntax for defining the code chunks and
|
||||
documentation chunks. The format is detected based on the file extension, but
|
||||
you can also set it manually using the `informat` parameter.
|
||||
documentation chunks. You can also weave Jupyter notebooks. The format is detected based on the file extension, but you can also set it manually using the `informat` parameter.
|
||||
|
||||
The rules for autodetection are:
|
||||
|
||||
```
|
||||
ext == ".jl" && return "script"
|
||||
ext == ".jmd" && return "markdown"
|
||||
ext == ".ipynb" && return "notebook"
|
||||
return "noweb"
|
||||
```
|
||||
|
||||
|
||||
|
||||
## Noweb
|
||||
|
||||
### Code chunks
|
||||
|
|
@ -104,4 +102,4 @@ Are the rest of the document (between `@` and `<<>>=` lines and the first chunk
|
|||
|
||||
## Markdown
|
||||
|
||||
Markdown code chunks are defined using fenced code blocks. [See sample document:](https://github.com/mpastell/Weave.jl/blob/master/examples/gadfly_sample.jmd)
|
||||
Markdown code chunks are defined using fenced code blocks. [See sample document:](https://github.com/mpastell/Weave.jl/blob/master/examples/gadfly_md_sample.jmd)
|
||||
|
|
|
|||
|
|
@ -109,11 +109,6 @@ const postexecute_hooks = Function[]
|
|||
push_postexecute_hook(f::Function) = push!(postexecute_hooks, f)
|
||||
pop_postexecute_hook(f::Function) = splice!(postexecute_hooks, findfirst(postexecute_hooks, f))
|
||||
|
||||
|
||||
|
||||
export weave, list_out_formats, tangle, convert_doc,
|
||||
set_chunk_defaults, get_chunk_defaults, restore_chunk_defaults
|
||||
|
||||
include("config.jl")
|
||||
include("chunks.jl")
|
||||
include("display_methods.jl")
|
||||
|
|
@ -123,4 +118,7 @@ include("cache.jl")
|
|||
include("formatters.jl")
|
||||
include("pandoc.jl")
|
||||
include("writers.jl")
|
||||
|
||||
export weave, list_out_formats, tangle, convert_doc,
|
||||
set_chunk_defaults, get_chunk_defaults, restore_chunk_defaults
|
||||
end
|
||||
|
|
|
|||
|
|
@ -29,14 +29,23 @@ function detect_outformat(outfile::String)
|
|||
return "noweb"
|
||||
end
|
||||
|
||||
"""Convert Weave documents between different formats"""
|
||||
"""
|
||||
`convert_doc(infile::AbstractString, outfile::AbstractString; format = nothing)`
|
||||
|
||||
Convert Weave documents between different formats
|
||||
|
||||
* `infile` = Name of the input document
|
||||
* `outfile` = Name of the output document
|
||||
* `format` = Output format (optional). Detected from outfile extension, but can
|
||||
be set to `"script"`, `"markdown"`, `"notebook"` or `"noweb"`.
|
||||
"""
|
||||
function convert_doc(infile::AbstractString, outfile::AbstractString; format = nothing)
|
||||
doc = read_doc(infile)
|
||||
|
||||
if format == nothing
|
||||
format = detect_outformat(outfile)
|
||||
end
|
||||
|
||||
|
||||
converted = convert_doc(doc, output_formats[format])
|
||||
|
||||
open(outfile, "w") do f
|
||||
|
|
|
|||
Loading…
Reference in New Issue